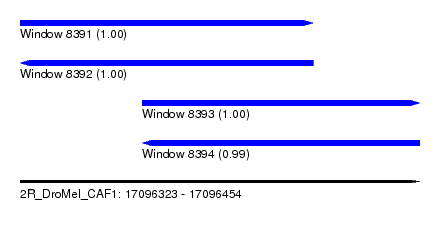
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,096,323 – 17,096,454 |
| Length | 131 |
| Max. P | 0.998835 |

| Location | 17,096,323 – 17,096,419 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -9.90 |
| Energy contribution | -11.68 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17096323 96 + 20766785 AGUCAUAAAAUCACUGCACAAAAUGCACUCAAUUGCACUUACAGAUGCAGAUACUGUAUCUGUAUC-----------------------UGCAUCUGCAUCCUCAUCUGGUUGCUGUCA .(.((...(((((..........((((......))))....(((((((((((((.......)))))-----------------------))))))))..........)))))..)).). ( -27.60) >DroSec_CAF1 33564 82 + 1 AGUCAUAAAAUCACUGCACAAAAUGCACUCAAUUGCACUUACAGAUACAGAUACUGUAUCUGUAUC-----------------------UGCAUCUGUAUCCGCA-------------- ..............((((.....))))..((..((((..(((((((((((...)))))))))))..-----------------------))))..))........-------------- ( -22.90) >DroSim_CAF1 33791 96 + 1 AGUCAUAAAAUCACUGCACAAAAUGCACUCAAUUGCACUUACAGAUACAGAUACUGUAUCUGUAUC-----------------------UGCAUCUGUAUCCGCAUCUGGUCGCUGUCA ............((.((((...((((...((..((((..(((((((((((...)))))))))))..-----------------------))))..)).....))))...)).)).)).. ( -28.00) >DroEre_CAF1 34155 119 + 1 AGUCGUAAAAUCACUGCACAAAACGCACUCAAUUGCACUUACAGAUACAGAUACUGUAUCUGCAUCAGCAUCGCCUCAGCCUCAGCAUUAGCAUCUGCAUCUGCAUAUGGUUUCUGUCA .((.((......)).))(((((((((.......(((.....(((((((((...))))))))).....)))..((....))....)).....(((.(((....))).))))))).))).. ( -23.10) >DroYak_CAF1 36117 108 + 1 AGUCGUAAAAUCACUGCAUGAAACGCACUCAAUUGCACUUACAUAUACAGAUACAGAUACAGACUCAG-----------AUACUGUAUCUGCAUCUGCGUCUGCAUCUGGUUGCUGUCA ............((.(((......(((......)))...........(((((.((((..((((..(((-----------(((...))))))..))))..)))).)))))..))).)).. ( -28.40) >DroPer_CAF1 67083 84 + 1 AGUCAUAAAAUCAAUGCACAAAUUGCAU-GAAAUGUCCGUACUUAU-----------AUCUGUAUC-----------------------UGCAUCAGAGUCUGUAUCUGUUUGAUUUCA .......(((((((((((.....)))))-...((((..((((....-----------....)))).-----------------------.))))((((.......))))..)))))).. ( -13.50) >consensus AGUCAUAAAAUCACUGCACAAAAUGCACUCAAUUGCACUUACAGAUACAGAUACUGUAUCUGUAUC_______________________UGCAUCUGCAUCCGCAUCUGGUUGCUGUCA ..............(((.......(((......)))...(((((((((((...)))))))))))..........................))).......................... ( -9.90 = -11.68 + 1.78)

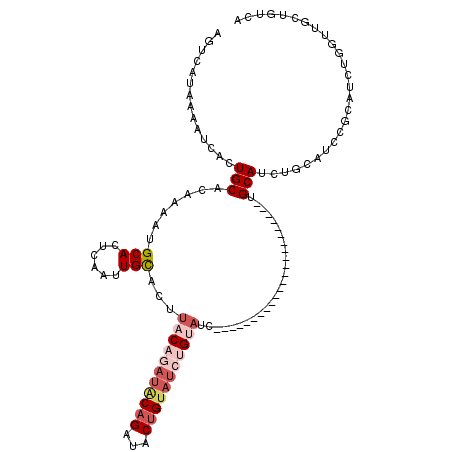
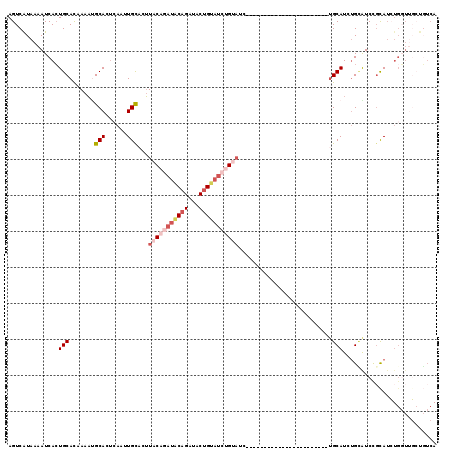
| Location | 17,096,323 – 17,096,419 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.12 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -16.50 |
| Energy contribution | -18.17 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17096323 96 - 20766785 UGACAGCAACCAGAUGAGGAUGCAGAUGCA-----------------------GAUACAGAUACAGUAUCUGCAUCUGUAAGUGCAAUUGAGUGCAUUUUGUGCAGUGAUUUUAUGACU .....(((.((......)).((((((((((-----------------------(((((.......)))))))))))))))((((((......))))))...)))............... ( -32.40) >DroSec_CAF1 33564 82 - 1 --------------UGCGGAUACAGAUGCA-----------------------GAUACAGAUACAGUAUCUGUAUCUGUAAGUGCAAUUGAGUGCAUUUUGUGCAGUGAUUUUAUGACU --------------((((..((((((((((-----------------------(((((.......)))))))))))))))..))))..(.(.(((((...))))).).).......... ( -30.50) >DroSim_CAF1 33791 96 - 1 UGACAGCGACCAGAUGCGGAUACAGAUGCA-----------------------GAUACAGAUACAGUAUCUGUAUCUGUAAGUGCAAUUGAGUGCAUUUUGUGCAGUGAUUUUAUGACU .....(((..(((.((((..((((((((((-----------------------(((((.......)))))))))))))))..)))).)))..)))........................ ( -33.40) >DroEre_CAF1 34155 119 - 1 UGACAGAAACCAUAUGCAGAUGCAGAUGCUAAUGCUGAGGCUGAGGCGAUGCUGAUGCAGAUACAGUAUCUGUAUCUGUAAGUGCAAUUGAGUGCGUUUUGUGCAGUGAUUUUACGACU ((((((((......((((((((((((((((..(((...(((.........)))...))).....)))))))))))))))).(..(......)..).)))))).)).............. ( -37.20) >DroYak_CAF1 36117 108 - 1 UGACAGCAACCAGAUGCAGACGCAGAUGCAGAUACAGUAU-----------CUGAGUCUGUAUCUGUAUCUGUAUAUGUAAGUGCAAUUGAGUGCGUUUCAUGCAGUGAUUUUACGACU .....(((..((.(((((((..(((((((((((.(((...-----------))).)))))))))))..))))))).))....))).....(((.(((.((((...))))....)))))) ( -38.10) >DroPer_CAF1 67083 84 - 1 UGAAAUCAAACAGAUACAGACUCUGAUGCA-----------------------GAUACAGAU-----------AUAAGUACGGACAUUUC-AUGCAAUUUGUGCAUUGAUUUUAUGACU (((((((((.((((.......)))).....-----------------------.........-----------....(((((((......-......))))))).)))))))))..... ( -13.50) >consensus UGACAGCAACCAGAUGCAGAUGCAGAUGCA_______________________GAUACAGAUACAGUAUCUGUAUCUGUAAGUGCAAUUGAGUGCAUUUUGUGCAGUGAUUUUAUGACU ..............(((((((((((((((....................................))))))))))))))).(((.((((.(.((((.....)))).))))).))).... (-16.50 = -18.17 + 1.67)
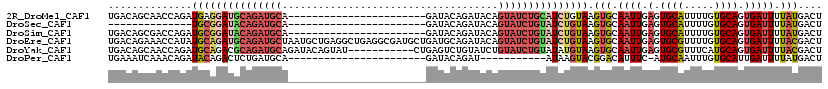
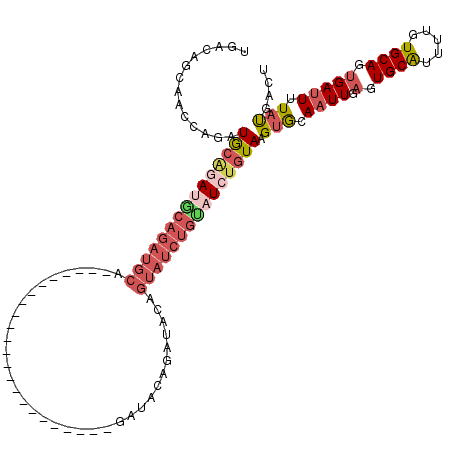
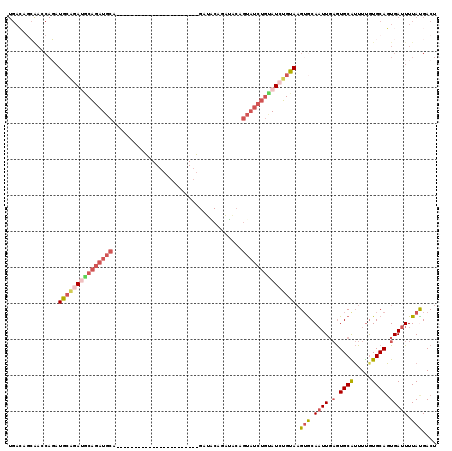
| Location | 17,096,363 – 17,096,454 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.60 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -10.32 |
| Energy contribution | -13.68 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17096363 91 + 20766785 ACAGAUGCAGAUAC-UGUAUCUGUAUC-----------------------UGCAUCUGCAUCCUCAUCUGGUUGCUGUCAAAUCUAGCUAUCGA--GUGCAGACUU---AAGGCAUUCAU .(((((((((((((-.......)))))-----------------------))))))))...........(((.((((.......)))).)))((--((((......---...)))))).. ( -32.70) >DroSim_CAF1 33831 91 + 1 ACAGAUACAGAUAC-UGUAUCUGUAUC-----------------------UGCAUCUGUAUCCGCAUCUGGUCGCUGUCAAAUCCAGCCAUCGA--GUGCAGACUU---AAGGCAUUCAU ((((((.(((((((-.......)))))-----------------------)).)))))).((.(((..((((.((((.......)))).)))).--.))).))...---........... ( -29.80) >DroEre_CAF1 34195 114 + 1 ACAGAUACAGAUAC-UGUAUCUGCAUCAGCAUCGCCUCAGCCUCAGCAUUAGCAUCUGCAUCUGCAUAUGGUUUCUGUCAAAUCCAGCAAUCGA--GUGCAGACUU---AAGGCAUUCAU .(((((((((...)-))))))))..........((((..((....((....))....)).(((((((.(((((.(((.......))).))))).--)))))))...---.))))...... ( -34.50) >DroYak_CAF1 36157 103 + 1 ACAUAUACAGAUAC-AGAUACAGACUCAG-----------AUACUGUAUCUGCAUCUGCGUCUGCAUCUGGUUGCUGUCAAAUCCAGCAAUCGA--GUGCAGACUU---AAGGCAUUCAU .......(((((.(-((((((((......-----------...))))))))).))))).(((((((..(((((((((.......))))))))).--.)))))))..---........... ( -43.20) >DroAna_CAF1 45490 79 + 1 ACAAAUGCAGAUACGUGUAUCUGUAUC-----------------------UGU------------------CGGCUGACAAAUUCCAAAUUCCACAUGGCAGAUUGUGGUAGAUACUUAU ....(((((((((....))))))))).-----------------------(((------------------(....))))...........(((((........)))))........... ( -17.90) >consensus ACAGAUACAGAUAC_UGUAUCUGUAUC_______________________UGCAUCUGCAUCCGCAUCUGGUUGCUGUCAAAUCCAGCAAUCGA__GUGCAGACUU___AAGGCAUUCAU ...((((((((((....))))))))))........................((.(((((((.......((((.((((.......)))).))))...)))))))(((...)))))...... (-10.32 = -13.68 + 3.36)

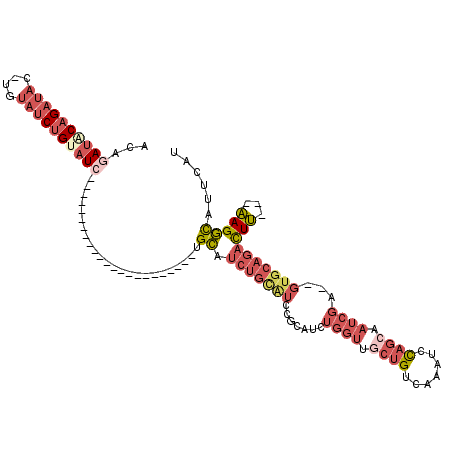
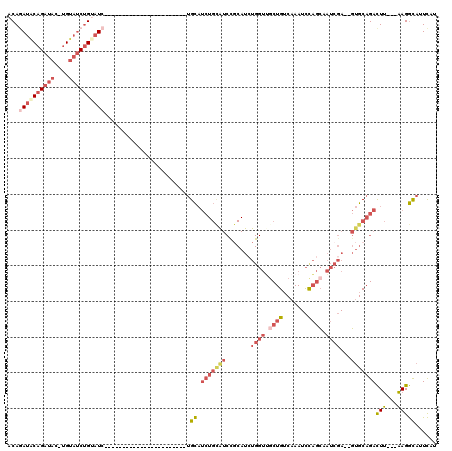
| Location | 17,096,363 – 17,096,454 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.60 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -9.89 |
| Energy contribution | -12.45 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.29 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17096363 91 - 20766785 AUGAAUGCCUU---AAGUCUGCAC--UCGAUAGCUAGAUUUGACAGCAACCAGAUGAGGAUGCAGAUGCA-----------------------GAUACAGAUACA-GUAUCUGCAUCUGU .....(((.((---((((((((..--......)).))))))))..))).((......))..(((((((((-----------------------(((((.......-)))))))))))))) ( -33.60) >DroSim_CAF1 33831 91 - 1 AUGAAUGCCUU---AAGUCUGCAC--UCGAUGGCUGGAUUUGACAGCGACCAGAUGCGGAUACAGAUGCA-----------------------GAUACAGAUACA-GUAUCUGUAUCUGU ...........---..((((((((--(.(.(.((((.......)))).)).)).)))))))(((((((((-----------------------(((((.......-)))))))))))))) ( -36.10) >DroEre_CAF1 34195 114 - 1 AUGAAUGCCUU---AAGUCUGCAC--UCGAUUGCUGGAUUUGACAGAAACCAUAUGCAGAUGCAGAUGCUAAUGCUGAGGCUGAGGCGAUGCUGAUGCAGAUACA-GUAUCUGUAUCUGU .....((((((---(.(((((((.--.......(((.......)))........)))))))((....((....))....)))))))))........(((((((((-(...)))))))))) ( -36.79) >DroYak_CAF1 36157 103 - 1 AUGAAUGCCUU---AAGUCUGCAC--UCGAUUGCUGGAUUUGACAGCAACCAGAUGCAGACGCAGAUGCAGAUACAGUAU-----------CUGAGUCUGUAUCU-GUAUCUGUAUAUGU ...........---..((((((((--(.(.((((((.......))))))).)).)))))))((((((((((((((((...-----------......))))))))-))))))))...... ( -45.80) >DroAna_CAF1 45490 79 - 1 AUAAGUAUCUACCACAAUCUGCCAUGUGGAAUUUGGAAUUUGUCAGCCG------------------ACA-----------------------GAUACAGAUACACGUAUCUGCAUUUGU ....((((((.(((((........)))))......(.(((((((....)------------------)))-----------------------))).)))))))................ ( -19.10) >consensus AUGAAUGCCUU___AAGUCUGCAC__UCGAUUGCUGGAUUUGACAGCAACCAGAUGCAGAUGCAGAUGCA_______________________GAUACAGAUACA_GUAUCUGUAUCUGU ..............((((((((..........)).))))))..............((((((((((((((.....................................)))))))))))))) ( -9.89 = -12.45 + 2.56)

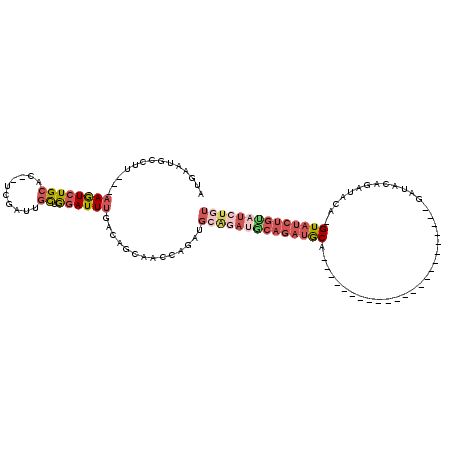
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:18 2006