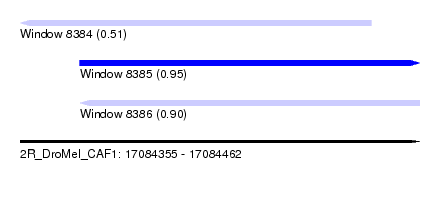
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,084,355 – 17,084,462 |
| Length | 107 |
| Max. P | 0.946024 |

| Location | 17,084,355 – 17,084,449 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17084355 94 - 20766785 GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCAGGCCAAUCCGCUUUGCCCGCCGGCACCAAAUUGCACAGCAACAGCAGCA ((..........(((((((......))))))).((.((((((((.(((.((......))))).).))))......((((...))))))))))). ( -26.00) >DroVir_CAF1 31682 88 - 1 GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUAUCGCCUGCGGCCAAUCCGUUUUGCCCCCUGGCCACAAAUUGCACAGCGGC------A ............(((((((......))))))).......(((((.(((((....(.....)....))))).))....((...)))))------. ( -20.90) >DroGri_CAF1 28154 88 - 1 CCACAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUAUGGCCGGCGGCCAAUCCGUUUUGCCCCUUGGCCACAAAUUGCACAGCGGC------A ............(((((((......))))))).....(((((...)))))..(((((.(((...(((....)))...))).))))).------. ( -24.50) >DroSec_CAF1 21722 94 - 1 GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCAGGCCAAUCCGCUUUGCCCGCCGGCACCAAAUUGCACAGCAACAGCAGCA ((..........(((((((......))))))).((.((((((((.(((.((......))))).).))))......((((...))))))))))). ( -26.00) >DroSim_CAF1 21893 94 - 1 GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCGGGCCAAUCCGCUUUGCCCGCCGGCACCAAAUUGCACAGCAACAGCAGCA ((..........(((((((......))))))).((.((((((((((((.((......))))))).))))......((((...))))))))))). ( -32.10) >DroYak_CAF1 23713 85 - 1 GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCAGGCCAAUCCGCUUUGCCCGCCGGCAACAAAUUGCACAGCA--------- (((.........(((((((......))))))).((.(((((.((((((......)).))))..)))))))......)))......--------- ( -23.50) >consensus GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCAGGCCAAUCCGCUUUGCCCGCCGGCAACAAAUUGCACAGCAAC______A (((.........(((((((......))))))).......(((((.(((.((......))))).).)))).......)))............... (-19.44 = -19.22 + -0.22)
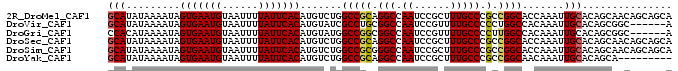
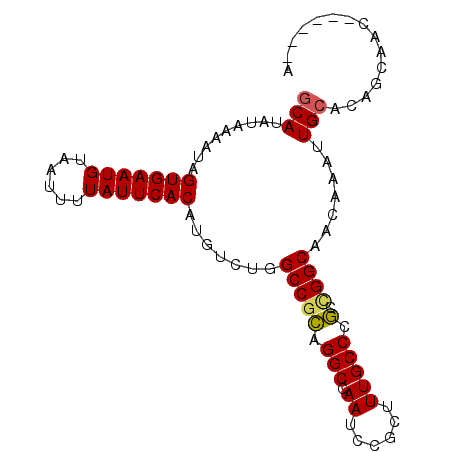
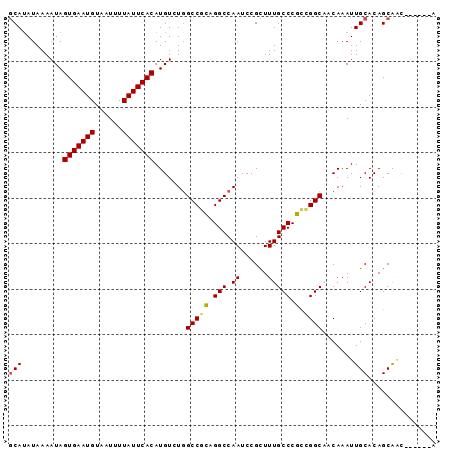
| Location | 17,084,371 – 17,084,462 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
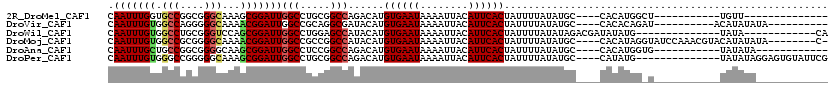
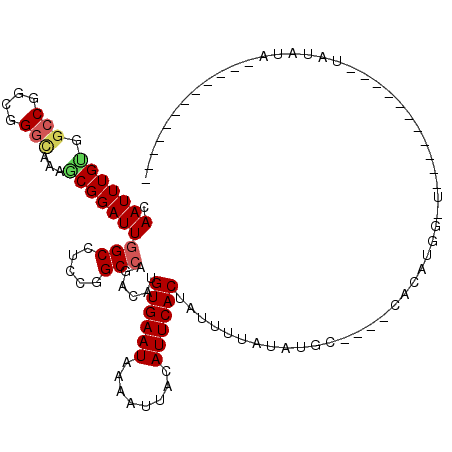
>2R_DroMel_CAF1 17084371 91 + 20766785 CAAUUUGGUGCCGGCGGGCAAAGCGGAUUGGCCUGCGGCCAGACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUGC----CACAUGGCU-----------UGUU-------------- (((((((.((((....))))...)))))))(((((.(((.......((((((........))))))..........))----).)).))).-----------....-------------- ( -26.03) >DroVir_CAF1 31692 96 + 1 CAAUUUGUGGCCAGGGGGCAAAACGGAUUGGCCGCAGGCGAUACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUGC----CACACAGAU----------ACAUAUAUA---------- .....(((((((((..(......)...)))))))))(((((((...((((((........)))))).....))).)))----)........----------.........---------- ( -24.30) >DroWil_CAF1 63947 94 + 1 CAAUUUGUGGCCUGCGGGUCCAGCGGAUUGGCCUGGAGCCAUACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUAGACGAUAUAUG--------------UAUA------------CA .....((((((.(.((((.((((....)))))))).)))))))...((((((........)))))).....((((((.....))))))--------------....------------.. ( -23.80) >DroMoj_CAF1 27499 107 + 1 CAAUUUGUGGCCGCGGGGCAAAACGGAUUGGCCGCCGGCCAUACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUGC----CACAUAGGUAUCCAAACGUACAUAUAUA--------C- .....((((((((.(.(((...........))).)))))))))...((((((........))))))........((((----(.....))))).................--------.- ( -28.00) >DroAna_CAF1 24002 93 + 1 CAAUUUGCUGCCGGCGGGGCAAGCGGAUUGGCCUCCGGCCAGACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUGC----CACAUGGUG-----------UAUAUA------------ .....(((((((((.(((.(((.....))).))))))).))).)).((((((........)))))).....(((((((----(....))))-----------))))..------------ ( -30.20) >DroPer_CAF1 56005 102 + 1 CAAUUUGUGGGCCGGGGGCAAAGCGGAUUGGCCUGCGGCCAGACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUGC----CAUAUG--------------UAUAUAGGAGUGUAUUCG ((...(((.(((((.((((.((.....)).)))).)))))..)))..))........(((((((.((((..((((((.----..))))--------------)).))))))))))).... ( -32.70) >consensus CAAUUUGUGGCCGGCGGGCAAAGCGGAUUGGCCUCCGGCCAGACAUGUGAAUAAAAUUACAUUCACUAUUUUAUAUGC____CACAUGG_U___________UAUAUA____________ .(((((((.(((....)))...)))))))(((.....)))......((((((........))))))...................................................... (-15.40 = -15.68 + 0.28)
| Location | 17,084,371 – 17,084,462 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.17 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
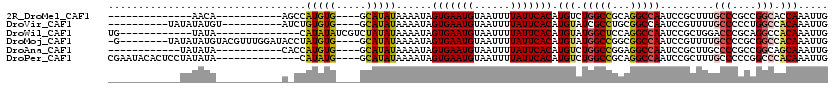
>2R_DroMel_CAF1 17084371 91 - 20766785 --------------AACA-----------AGCCAUGUG----GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCAGGCCAAUCCGCUUUGCCCGCCGGCACCAAAUUG --------------....-----------.(((.((((----((.......((.(((((((......))))))))).....))))))((((((......)))...))))))......... ( -26.00) >DroVir_CAF1 31692 96 - 1 ----------UAUAUAUGU----------AUCUGUGUG----GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUAUCGCCUGCGGCCAAUCCGUUUUGCCCCCUGGCCACAAAUUG ----------.........----------.....((((----((..........(((((((......))))))).......((..((((.....))))...))......))))))..... ( -23.30) >DroWil_CAF1 63947 94 - 1 UG------------UAUA--------------CAUAUAUCGUCUAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUAUGGCUCCAGGCCAAUCCGCUGGACCCGCAGGCCACAAAUUG ((------------((((--------------(((((((.....))))).....(((((((......))))))).)))))((((.(.(((((......))).)).)..)))))))..... ( -21.00) >DroMoj_CAF1 27499 107 - 1 -G--------UAUAUAUGUACGUUUGGAUACCUAUGUG----GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUAUGGCCGGCGGCCAAUCCGUUUUGCCCCGCGGCCACAAAUUG -.--------..(((((((((((..((...)).)))).----))))))).....(((((((......))))))).....((((((.(((.(((......)))..)))))))))....... ( -31.40) >DroAna_CAF1 24002 93 - 1 ------------UAUAUA-----------CACCAUGUG----GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGGAGGCCAAUCCGCUUGCCCCGCCGGCAGCAAAUUG ------------......-----------(((...)))----((..........(((((((......))))))).((.(((((.((.(((..........))))))))))))))...... ( -25.00) >DroPer_CAF1 56005 102 - 1 CGAAUACACUCCUAUAUA--------------CAUAUG----GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUCUGGCCGCAGGCCAAUCCGCUUUGCCCCCGGCCCACAAAUUG ...........(((((..--------------..))))----)...........(((((((......))))))).(((..(((((..(((.((......)))))..))))).)))..... ( -23.10) >consensus ____________UAUAUA___________A_CCAUGUG____GCAUAUAAAAUAGUGAAUGUAAUUUUAUUCACAUGUAUGGCCGCAGGCCAAUCCGCUUUGCCCCCCGGCCACAAAUUG .................................(((((.....)))))......(((((((......))))))).(((.(((((...))))).........(((....))).)))..... (-14.22 = -14.17 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:10 2006