
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,082,619 – 17,082,722 |
| Length | 103 |
| Max. P | 0.833702 |

| Location | 17,082,619 – 17,082,722 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.66 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17082619 103 - 20766785 UAGGA----AUCAUUAUGCAGUUAAUGACAAAUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUCCAGAAGAGGCCCCUUU--CCGUUCCACUUUUGCC ..(((----(((((((.(((((...((((....))))...))))))))))).....(((.((((.....))))))).(.((((....))))--.).))))......... ( -27.80) >DroPse_CAF1 52206 99 - 1 UAGGAAUGAAUCAUUAUGCAGUUAAUGACAAGUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUCCUG----GCUCCAUCACCCGGACCCACC------ ..((......((((((.(((((...((((....))))...)))))))))))....((((.((((.....))))))))(----(.(((.......))).)).))------ ( -27.90) >DroSec_CAF1 20051 105 - 1 UAGGA----AUCAUUAUGCAGUUAAUGACAAAUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUGCAGAAGAGCCCCCUCCCCCCGUCCCACUUUUCCC ..(((----(((((((.(((((...((((....))))...))))))))))).....(((((((.(((......))).)).(((....)))....))))).....)))). ( -26.20) >DroSim_CAF1 20153 105 - 1 UAGGA----AUCAUUAUGCAGUUAAUGACAAAUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUGCAGAAGAGCCACCUCCCCCCUUCCCACUUUUCCC ..(((----(......((((((...((((....))))((.((((((...((((......))))))))))))))))))((((.............))))......)))). ( -25.62) >DroEre_CAF1 20509 94 - 1 UAGGA----AUCAUUAUGCAGUUAAUGACAAAUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUCCC-A----------GCCCCUGCCCCACUUUCCCC ..(((----(((((((.(((((...((((....))))...))))))))))).....(((.((((.....))))))).-.----------..............)))).. ( -24.30) >DroYak_CAF1 22020 95 - 1 UAGGA----AUCAUUAUGCAGUUAAUGACAAAUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUCCAGA----------UCCCCUGCCCCACUUUCCUC .((((----(((((((.(((((...((((....))))...))))))))))).....(((.((((.....)))))))...----------..............))))). ( -26.10) >consensus UAGGA____AUCAUUAUGCAGUUAAUGACAAAUGUCACGCAUUGCUAAUGACAAAAGGAUGUCGGCAAUCGACUCCAGA___GCCCCCUCCCCCCGUCCCACUUUUCCC ..((......((((((.(((((...((((....))))...))))))))))).....(((.((((.....)))))))......................))......... (-21.40 = -21.73 + 0.33)

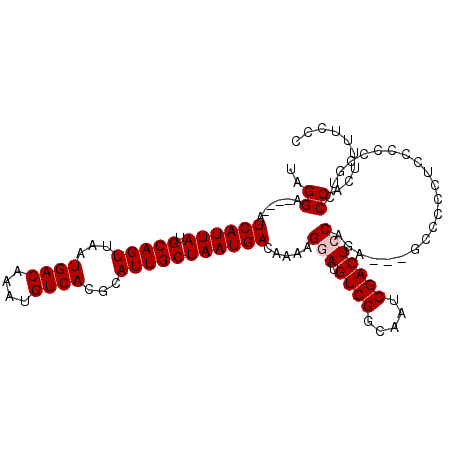
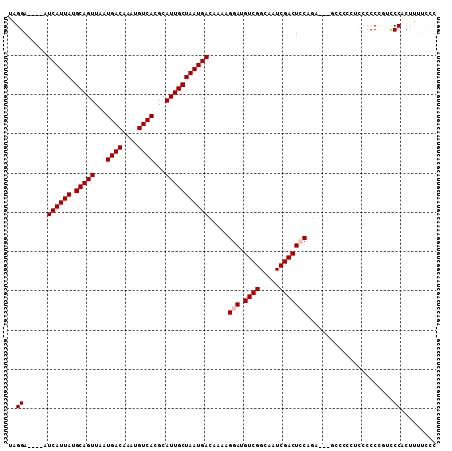
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:08 2006