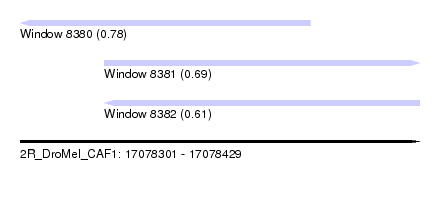
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,078,301 – 17,078,429 |
| Length | 128 |
| Max. P | 0.779821 |

| Location | 17,078,301 – 17,078,394 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -14.99 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17078301 93 - 20766785 UGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCGUAAACUGA--------------AAACUUUUC-AUUUGCAAAAUGCCCCGA .(((((.((((...))))((((((...((((((....))).)))...))))))....(((((.(((--------------(.....)))-))))))..)))))..... ( -24.10) >DroVir_CAF1 23820 81 - 1 UGCAUUGGAUCAACGAUCACCAUUGACGGUGGAGUCCUCCUGCCCCAAACGGUGAGUA--AAUCUCA-------------AUCC--------AAGCAAAA----CAAA (((.((((((......(((((.((...((((((....))).)))...)).)))))...--.......-------------))))--------)))))...----.... ( -21.49) >DroSec_CAF1 15774 93 - 1 UGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCGUAAACUGA--------------AAACUUUUC-AUUUGCAAAAUGCCCCGA .(((((.((((...))))((((((...((((((....))).)))...))))))....(((((.(((--------------(.....)))-))))))..)))))..... ( -24.10) >DroSim_CAF1 15882 92 - 1 UGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCA-ACGGUAAGCAUAAACUGA--------------AAACUUUUC-AUUUGCAAAAUGCCCCGA .(((((.((((...))))(((((((..((((((....))).))).))-)))))..(((.....(((--------------(.....)))-)..)))..)))))..... ( -24.00) >DroYak_CAF1 17677 107 - 1 UGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCGUAAAUUGAAAACUAAAAACUGAAAACUUUUC-AUUUGCAAAAUGCCCCAA .(((((.((((...))))((((((...((((((....))).)))...))))))....((((((.(((((...............)))))-))))))..)))))..... ( -24.46) >DroMoj_CAF1 18792 89 - 1 UGCAUUGGAUCAACGAUCACCAUUGACGGUGGAGUCCUCGUGUCCCAAGCGGUAAGUA--AAUUCCA-------------ACGCUUUUCUAGCAGCAAAG----CAAA (((.((((..((.((((((((......))))).....)))))..))))((((......--....)).-------------..(((.....))).))...)----)).. ( -20.90) >consensus UGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCAUAAACUGA______________AAACUUUUC_AUUUGCAAAAUGCCCCAA (((....((((...))))((((((...((((((....))).)))...)))))).........................................)))........... (-14.99 = -15.38 + 0.39)

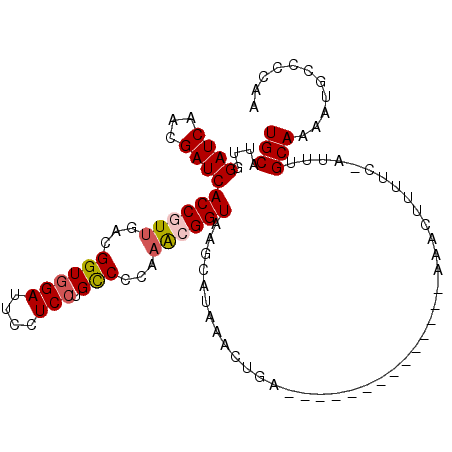

| Location | 17,078,328 – 17,078,429 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17078328 101 + 20766785 UCAGUUUA---CGCUUACCGUUUGGGGCAGGAGGAAUCCACCGUCAACGGUGAUCGUUGAUCCAAUGCAACAAACGGAGUUCUGUUU-UUGGGAGG-AAAG---CAAAG ........---.((((.(((((((..((((((..(((.(((((....)))))...)))..)))..)))..)))))))..((((.(..-...).)))-))))---).... ( -32.30) >DroPse_CAF1 47832 96 + 1 U----------UGCUCACCGUUUGGGGCAGGAGGAAUCCACCGUCAACGGUGAUCGUUGAUCCAAUGCAACAAACGGAGUCCUGUGC-GGGGGAGAGAAAU--CGAAAG .----------..(((.(((((((..((((((..(((.(((((....)))))...)))..)))..)))..)))))))..((((....-..)))))))....--...... ( -33.50) >DroSec_CAF1 15801 102 + 1 UCAGUUUA---CGCUUACCGUUUGGGGCAGGAGGAAUCCACCGUCAACGGUGAUCGUUGAUCCAAUGCAACAAACGGAGUUCUGUUUUUUGGGAGG-AAAG---CAAAG ........---.((((.(((((((..((((((..(((.(((((....)))))...)))..)))..)))..)))))))..((((.((....)).)))-))))---).... ( -33.00) >DroSim_CAF1 15909 100 + 1 UCAGUUUA---UGCUUACCGU-UGGGGCAGGAGGAAUCCACCGUCAACGGUGAUCGUUGAUCCAAUGCAACAAACGGAGUUCUGUUU-UUGGAAGG-AAAG---CAAUG ........---(((((.((((-(((((..(((....))).))(((((((.....)))))))))))).(((.((((((....))))))-)))...))-.)))---))... ( -30.20) >DroWil_CAF1 55848 97 + 1 UGAGUUCAAUUCACUUACCGUUUGGGGCAAGAGGAAUCCACCGUCAACGGUGAUCUUUGAUCCAAUGCAACAAAUGGAGUUCUGUGG---AGCAGG---------AAAC (((((.......)))))(((((((..(((...(((.(((((((....)))))......)))))..)))..)))))))..(((((...---..))))---------)... ( -27.90) >DroAna_CAF1 17743 101 + 1 ---UUUUC---GACUUACCGUUUGGGGCAGGAGGAAUCCACCGUCAACGGCGAUCGUUGAUCCAAUGCAACAAACGGAGUUCUGUUU-UUGGUAAG-AGAUUUGGAGAG ---(((((---((....(((((((..((((((....)))...(((((((.....)))))))....)))..)))))))...(((.((.-.....)).-))).))))))). ( -31.30) >consensus UCAGUUUA___CGCUUACCGUUUGGGGCAGGAGGAAUCCACCGUCAACGGUGAUCGUUGAUCCAAUGCAACAAACGGAGUUCUGUUU_UUGGGAGG_AAAG___CAAAG .................(((((((..((((((..(((.(((((....)))))...)))..)))..)))..)))))))................................ (-24.45 = -24.98 + 0.53)

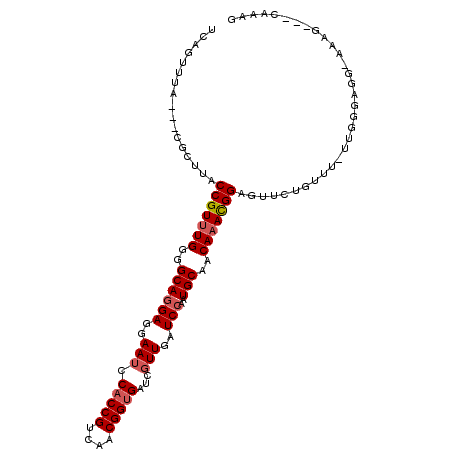
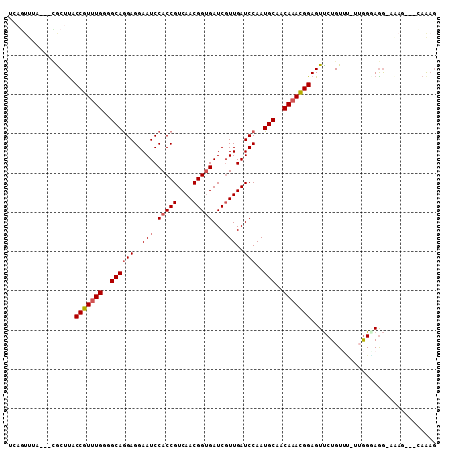
| Location | 17,078,328 – 17,078,429 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17078328 101 - 20766785 CUUUG---CUUU-CCUCCCAA-AAACAGAACUCCGUUUGUUGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCG---UAAACUGA ....(---(((.-((..((((-.((((((......))))))...)))).((((((.....)))))).((((((....))).)))......)).)))).---........ ( -24.60) >DroPse_CAF1 47832 96 - 1 CUUUCG--AUUUCUCUCCCCC-GCACAGGACUCCGUUUGUUGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUGAGCA----------A ......--....(((....((-.....))...(((((((..(((..(((.......((((((....))))))..)))...)))..))))))).)))..----------. ( -24.80) >DroSec_CAF1 15801 102 - 1 CUUUG---CUUU-CCUCCCAAAAAACAGAACUCCGUUUGUUGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCG---UAAACUGA ....(---(((.-((..((((..((((((......))))))...)))).((((((.....)))))).((((((....))).)))......)).)))).---........ ( -25.00) >DroSim_CAF1 15909 100 - 1 CAUUG---CUUU-CCUUCCAA-AAACAGAACUCCGUUUGUUGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCA-ACGGUAAGCA---UAAACUGA ...((---(((.-...(((((-.((((((......))))))...))))).........(((((((..((((((....))).))).))-))))))))))---........ ( -27.20) >DroWil_CAF1 55848 97 - 1 GUUU---------CCUGCU---CCACAGAACUCCAUUUGUUGCAUUGGAUCAAAGAUCACCGUUGACGGUGGAUUCCUCUUGCCCCAAACGGUAAGUGAAUUGAACUCA ...(---------(((((.---..(((((......))))).)))..)))((((...((((((....)))))).(((..((((((......)))))).)))))))..... ( -24.80) >DroAna_CAF1 17743 101 - 1 CUCUCCAAAUCU-CUUACCAA-AAACAGAACUCCGUUUGUUGCAUUGGAUCAACGAUCGCCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGUC---GAAAA--- .........((.-((((((..-.((((((......))))))...((((.((((((.....)))))).((((((....))).)))))))..))))))..---))...--- ( -25.80) >consensus CUUUG___CUUU_CCUCCCAA_AAACAGAACUCCGUUUGUUGCAUUGGAUCAACGAUCACCGUUGACGGUGGAUUCCUCCUGCCCCAAACGGUAAGCG___UAAACUGA ................................(((((((..(((..(((.......((((((....))))))..)))...)))..)))))))................. (-20.69 = -20.88 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:07 2006