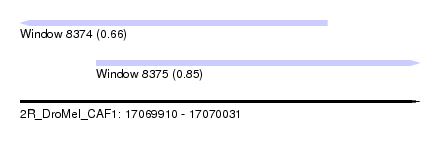
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,069,910 – 17,070,031 |
| Length | 121 |
| Max. P | 0.852183 |

| Location | 17,069,910 – 17,070,003 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -18.80 |
| Energy contribution | -19.88 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
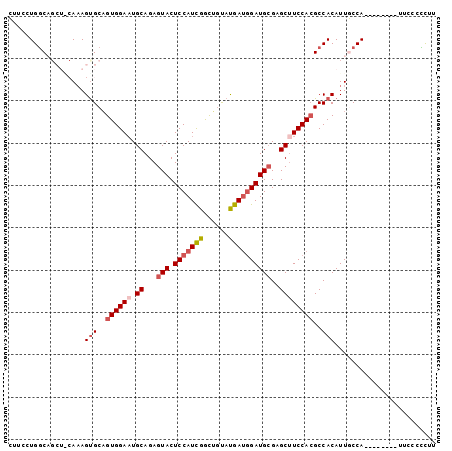
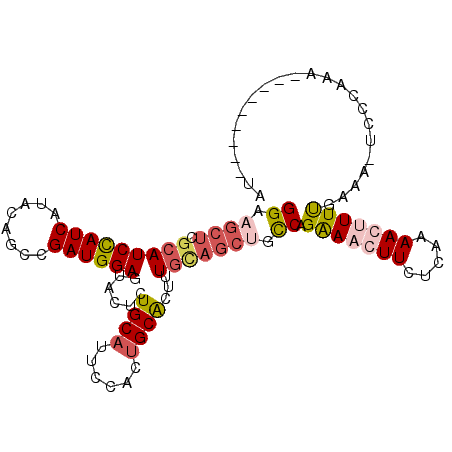
>2R_DroMel_CAF1 17069910 93 - 20766785 CAUCCUGGCAGCUACAAAGUGCAGUGGAAUGCAGAGUACUCCAUCGAUUGUAUGAUGGAUGCGAGCUUCCACGCCACAUUGCCA--------UUCCUCUUU .....(((((((........)).((((((.((...(((.(((((((......))))))))))..)))))))).......)))))--------......... ( -29.90) >DroSec_CAF1 7536 87 - 1 CUUCCUGGC------AAAGUGCAGUGGAAUGCAGAGUACUCCAUUGGCUAUAUGACGGAUGCGAGCUUCCACGCCACAUUGCCA--------UUCCCCCUU .....((((------((.(((..((((((.((...(((.(((.(((......))).))))))..))))))))..))).))))))--------......... ( -27.40) >DroSim_CAF1 7518 87 - 1 CUUCCUGGC------AAAGUGCAGUGGAAUGCAGAGUACUCCAUCGGCUGUAUGAUAGAUGGGAGCUUCCACGCCACAUUGCCA--------UUCCCCCUU .....((((------((.(((..((((((.((((....))(((((..(.....)...)))))..))))))))..))).))))))--------......... ( -28.10) >DroEre_CAF1 7592 101 - 1 GUUUCUGACAGCUGCAAAGCGCUGUGGAAUGCAGAGUACUCCAUCGGCAGCAUGAUGGAUGCGAGCAUCCACGCCACAUUUCCAUUGCCGCAUUCCCUCCU .......(((((.((...)))))))(((((((.((........))(((((...((((((((....)))))).........))..))))))))))))..... ( -30.70) >DroYak_CAF1 8906 93 - 1 GUUUCUGGCAGCUGCACAGUGCAGUGGAUUGCAGAGUACUCCAUCGGCAGUACGAUGGAUGCGAGCUUCCAGGCCACAUUUCCA--------UUCCCUCUU .....((((.(((((.....)))))(((..((...(((.(((((((......))))))))))..)).)))..))))........--------......... ( -31.90) >consensus CUUCCUGGCAGCU_CAAAGUGCAGUGGAAUGCAGAGUACUCCAUCGGCUGUAUGAUGGAUGCGAGCUUCCACGCCACAUUGCCA________UUCCCCCUU ..................(((..((((((.((...(((.(((((((......))))))))))..))))))))..)))........................ (-18.80 = -19.88 + 1.08)


| Location | 17,069,933 – 17,070,031 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -13.95 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
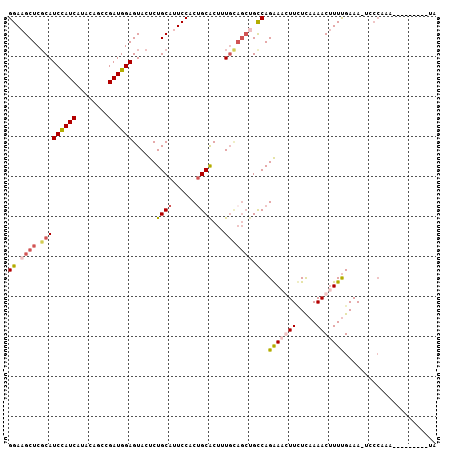
>2R_DroMel_CAF1 17069933 98 + 20766785 GGAAGCUCGCAUCCAUCAUACAAUCGAUGGAGUACUCUGCAUUCCACUGCACUUUGUAGCUGCCAGGAUGUUCUUAAAACUUUUGAAAAUACCAAA---------UA ((.((((.(((((((((........))))))......((((......))))...))))))).)).((.((((.((((.....)))).))))))...---------.. ( -22.50) >DroSim_CAF1 7541 95 + 1 GGAAGCUCCCAUCUAUCAUACAGCCGAUGGAGUACUCUGCAUUCCACUGCACUUU------GCCAGGAAGUUCUUAAAACUUUUGAAAGUCCCAAAUU------ACA ((((((.....((((((........)))))).......)).))))...((.....------))..(((..(((...........)))..)))......------... ( -17.80) >DroEre_CAF1 7623 107 + 1 GGAUGCUCGCAUCCAUCAUGCUGCCGAUGGAGUACUCUGCAUUCCACAGCGCUUUGCAGCUGUCAGAAACUUUUCAAAACCUUUGAAACUCCCACAUUUGCGGUAUA (((((....)))))...(((((((.((((((((..((((......(((((........))))))))).....(((((.....))))))))))...))).))))))). ( -32.90) >DroYak_CAF1 8929 88 + 1 GGAAGCUCGCAUCCAUCGUACUGCCGAUGGAGUACUCUGCAAUCCACUGCACUGUGCAGCUGCCAGAAACUUGUCAAAACUUUCGAAA------------------- ((.((((....(((((((......)))))))((((..((((......))))..)))))))).)).((((.((.....)).))))....------------------- ( -27.20) >consensus GGAAGCUCGCAUCCAUCAUACAGCCGAUGGAGUACUCUGCAUUCCACUGCACUUUGCAGCUGCCAGAAACUUCUCAAAACUUUUGAAA_UCCCAAA_________UA ((.((((.(((((((((........))))))......((((......))))...))))))).)).(((((((.....)))))))....................... (-13.95 = -15.82 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:00 2006