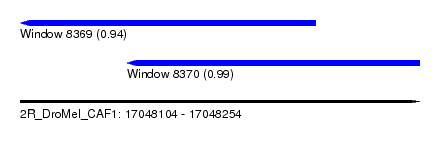
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,048,104 – 17,048,254 |
| Length | 150 |
| Max. P | 0.992323 |

| Location | 17,048,104 – 17,048,215 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.15 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -22.73 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17048104 111 - 20766785 AAACGAA-ACCUCCCAGUGGCAAACUUUG-----UUCAACCAUCA---GUAUGUUUGUACAGCUGGGAUAUGAAAAACACUAAGAUCUUUGUAGUGGUCGUACUUUUGGCCGCCAGUUCA ....(((-...(((((((.((((((.(((-----.........))---)...))))))...))))))).........................(((((((......)))))))...))). ( -28.90) >DroSec_CAF1 141592 114 - 1 AAGGGAA-ACAUCCCAGUUGCGAACUUUA-----UUCAACCAUUAGCUGUAUGUUUAUACAGCUGGGGUAUGAAAAACACUGAGAUCUUUGUACUGGCCGUACUUUUGGCCAGCAGUUCA ..((((.-...))))......(((((...-----(((((((.((((((((((....))))))))))))).))))...................(((((((......))))))).))))). ( -40.00) >DroSim_CAF1 150136 114 - 1 AAGCGAA-ACAUCCCAGUUGCGAACUUUA-----UUCAACCAUUAGCUGUAUGUUCAUACAGCUGGGGUAUGAAAAACACUGAGAUCUUUGUACUGGCCGUACUUUUGGCCAGCAGUUCA ..((((.-.........))))(((((...-----(((((((.((((((((((....))))))))))))).))))...................(((((((......))))))).))))). ( -37.20) >DroEre_CAF1 145698 116 - 1 CAUCGAACAGCUCCCAGUUGCGAGCUUCGAACAGCUCA-GCAUCA---GUAUGCUUGCACUGCUGGAAUAUGAGAAACACUGAAAUCUUGGUACUGGCCGUGCCUUUGGCUGCCUGCUCA .......(((((.(((((((((((((......)))))(-((((..---..))))).)))..))))).......................(((((.....)))))...)))))........ ( -31.70) >DroYak_CAF1 155186 112 - 1 UAGGAAACACAUCCCAGUUGCGUGAUGUG-----CUCAACCAUCA---GUAUGCUCGCACUGCUGGGAUAUGGAAAACAUGGAAAUCUUCGUACUGGCCGUGCUUUUGGCUGCCUGUUCA ((((...((.((((((((.(.((((((((-----((........)---))))).)))).).)))))))).))......(((((....)))))...(((((......))))).)))).... ( -33.50) >consensus AAGCGAA_ACAUCCCAGUUGCGAACUUUG_____UUCAACCAUCA___GUAUGUUUGUACAGCUGGGAUAUGAAAAACACUGAGAUCUUUGUACUGGCCGUACUUUUGGCCGCCAGUUCA ..........(((((((((((((((...........................)))))))..)))))))).((((....((.((....)).)).(((((((......)))))))...)))) (-22.73 = -21.77 + -0.96)

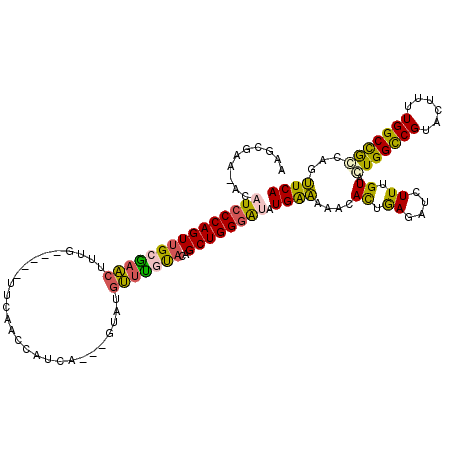
| Location | 17,048,144 – 17,048,254 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.32 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17048144 110 - 20766785 GCUCUCUUCUCUCCAACCGGCGAUA-AGCCAAUUCAUAAAAAACGAA-ACCUCCCAGUGGCAAACUUUG-----UUCAACCAUCA---GUAUGUUUGUACAGCUGGGAUAUGAAAAACAC ..................(((....-.)))..((((((.........-...(((((((.((((((.(((-----.........))---)...))))))...)))))))))))))...... ( -25.80) >DroSec_CAF1 141632 113 - 1 UCUCUCUUCUCUUCGAGAGGAGAUA-AGCCAAUUCAUAAAAAGGGAA-ACAUCCCAGUUGCGAACUUUA-----UUCAACCAUUAGCUGUAUGUUUAUACAGCUGGGGUAUGAAAAACAC ..(((((((((...)))))))))..-......(((.(((...((((.-...))))..))).))).....-----(((((((.((((((((((....))))))))))))).))))...... ( -34.50) >DroSim_CAF1 150176 113 - 1 GCUCUCUUCUCUCCGAGAGGCGAUA-AGCCAAUUCAUAAAAAGCGAA-ACAUCCCAGUUGCGAACUUUA-----UUCAACCAUUAGCUGUAUGUUCAUACAGCUGGGGUAUGAAAAACAC ..((((........))))(((....-.)))......((((...((.(-((......))).))...))))-----(((((((.((((((((((....))))))))))))).))))...... ( -29.00) >DroEre_CAF1 145738 112 - 1 GCGCUCUUCCCUUCG----GCCUUAUCACAAAUCCACAAACAUCGAACAGCUCCCAGUUGCGAGCUUCGAACAGCUCA-GCAUCA---GUAUGCUUGCACUGCUGGAAUAUGAGAAACAC ...(((.....((((----((.....................((((..(((((.(....).)))))))))...((..(-((((..---..))))).))...))))))....)))...... ( -24.40) >DroYak_CAF1 155226 108 - 1 GCUCUUUACUCUCCG----GCCUUAUCGCCAAUCCGUAAAUAGGAAACACAUCCCAGUUGCGUGAUGUG-----CUCAACCAUCA---GUAUGCUCGCACUGCUGGGAUAUGGAAAACAU ..............(----((......)))..((((((....(....)...(((((((.(.((((((((-----((........)---))))).)))).).)))))))))))))...... ( -32.80) >consensus GCUCUCUUCUCUCCGA__GGCGAUA_AGCCAAUUCAUAAAAAGCGAA_ACAUCCCAGUUGCGAACUUUG_____UUCAACCAUCA___GUAUGUUUGUACAGCUGGGAUAUGAAAAACAC ................................((((((.............((((((((((((((...........................)))))))..)))))))))))))...... (-14.64 = -14.32 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:56 2006