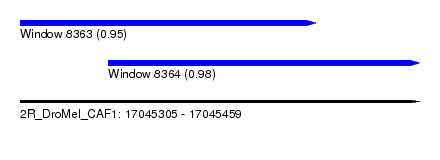
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,045,305 – 17,045,459 |
| Length | 154 |
| Max. P | 0.978817 |

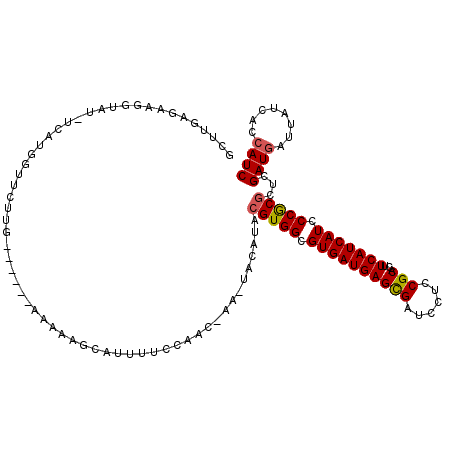
| Location | 17,045,305 – 17,045,419 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -19.02 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17045305 114 + 20766785 GCUUGAGAAGGUUUUUCGGGGUUCUUG------AAAAAGAAUUUUCCAACUAACUACAUACGUGGCGUGAUGAGCGAUCCUCCGCACUUCAUCAUCCCACGUUCGAUGAUUAUCACCAUC .........(((.....(((((((((.------...))))))..))).........(((((((((.((((((((((......)))...))))))).))))))...)))......)))... ( -32.10) >DroSec_CAF1 138685 111 + 1 GCUUGAGAAGGUAU-UAAUGGUUC--G------AAAGAGCAUUUUCCCACUAAAUACAUACGUGGCGUGAUGAGCGAUCCUCCGCAUUUCAUCAUCCCGCGCUCGAUGAUUAUCACCAUC .........(((..-((((.((((--.------...))))................(((.(((((.((((((((((......)))...))))))).)))))....)))))))..)))... ( -31.90) >DroSim_CAF1 147217 113 + 1 GCUUGAGAAGGUAU-UCAUGGUUCUUG------AAAGAGCAUUUUCCAACAAAAUACAUACGUGGCGUGAUGAGCGAUCCUCCGCAUUUCAUCAUCCCGCGCUCGAUGAUUAUCACCAUC .........((((.-((((.(((((..------..)))))....................(((((.((((((((((......)))...))))))).)))))....)))).))))...... ( -29.60) >DroEre_CAF1 142923 104 + 1 A--UCACAAGUUAU-UCAUAAUUCAUAAUUCCUGAAAAUCA-------------UACAUACGUGGCGUGAUGAGCGACCCGCCGCACGUCAUCAUCCCGCCCUUGAUGAUGAUCACCAUC .--...........-.................(((..((((-------------(.((...((((.((((((((((......)))...))))))).))))...)))))))..)))..... ( -24.10) >DroYak_CAF1 152431 109 + 1 ACUUCACAGGAUAU-UCACGAUUCUUG------UAAAAGCAUAUUCCAAC----UACAUACGUGGCGUGAUGAGUGACCCCCCGCACUUCAUCAUCCCGCGCUCGAUCACCAUCACCAUC ...((((((((...-.......)))))------)................----......(((((.((((((((((........))).))))))).)))))...)).............. ( -23.80) >consensus GCUUGAGAAGGUAU_UCAUGGUUCUUG______AAAAAGCAUUUUCCAAC_AA_UACAUACGUGGCGUGAUGAGCGAUCCUCCGCACUUCAUCAUCCCGCGCUCGAUGAUUAUCACCAUC ............................................................(((((.((((((((((......)))...))))))).)))))...((((........)))) (-19.02 = -19.10 + 0.08)


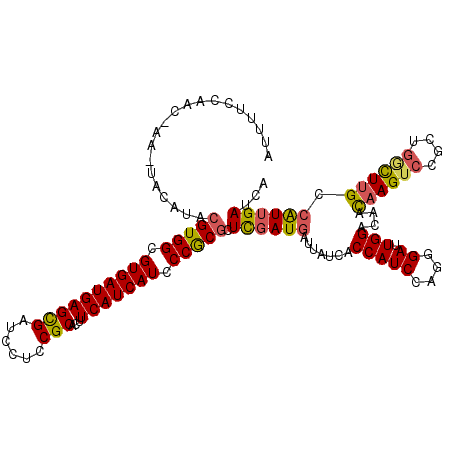
| Location | 17,045,339 – 17,045,459 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.00 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17045339 120 + 20766785 AUUUUCCAACUAACUACAUACGUGGCGUGAUGAGCGAUCCUCCGCACUUCAUCAUCCCACGUUCGAUGAUUAUCACCAUCCAGGGAUUGGAGAAUAAGUCCGCUGGCUUGCCAUUGAUCA ................(..((((((.((((((((((......)))...))))))).))))))..).((((((....((.(((((((((........))))).))))..))....)))))) ( -36.80) >DroSec_CAF1 138716 120 + 1 AUUUUCCCACUAAAUACAUACGUGGCGUGAUGAGCGAUCCUCCGCAUUUCAUCAUCCCGCGCUCGAUGAUUAUCACCAUCCAGGGAUUGGACAACAAGUCCGCUGGUUUGCCAUUGAUCA ....((((............(((((.((((((((((......)))...))))))).)))))...((((........))))..))))..((((.....))))..(((....)))....... ( -36.00) >DroSim_CAF1 147250 120 + 1 AUUUUCCAACAAAAUACAUACGUGGCGUGAUGAGCGAUCCUCCGCAUUUCAUCAUCCCGCGCUCGAUGAUUAUCACCAUCCAGGGAUUGGACAACAAGUCCGCUGGUUUGCCAUUGAUCA ....................(((((.((((((((((......)))...))))))).))))).....((((((....((.(((((((((........))))).))))..))....)))))) ( -35.10) >DroEre_CAF1 142960 107 + 1 A-------------UACAUACGUGGCGUGAUGAGCGACCCGCCGCACGUCAUCAUCCCGCCCUUGAUGAUGAUCACCAUCCAGGGAUUGGAAAACAAGUCUGCGGACUUGCCGUAGAUCA .-------------.......((((((((.((.(((...)))))))))))))..(((..((((.((((........)))).))))...)))......((((((((.....)))))))).. ( -38.70) >DroYak_CAF1 152464 116 + 1 AUAUUCCAAC----UACAUACGUGGCGUGAUGAGUGACCCCCCGCACUUCAUCAUCCCGCGCUCGAUCACCAUCACCAUCCAGGGAUUGGCUAAUAAGUCUGCAUCCUGGCCAUUGAUCA ..........----......(((((.((((((((((........))).))))))).)))))...(((((..........((((((...((((....))))....))))))....))))). ( -33.24) >consensus AUUUUCCAAC_AA_UACAUACGUGGCGUGAUGAGCGAUCCUCCGCACUUCAUCAUCCCGCGCUCGAUGAUUAUCACCAUCCAGGGAUUGGACAACAAGUCCGCUGGCUUGCCAUUGAUCA ....................(((((.((((((((((......)))...))))))).))))).((((((.......(((((....)).)))....((((((....)))))).))))))... (-27.04 = -27.00 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:50 2006