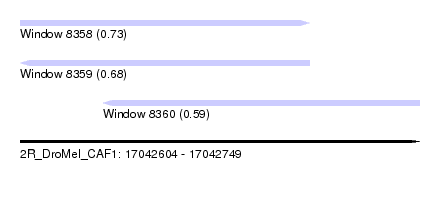
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,042,604 – 17,042,749 |
| Length | 145 |
| Max. P | 0.726002 |

| Location | 17,042,604 – 17,042,709 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
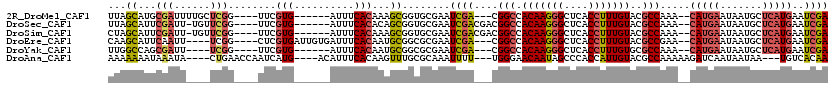
>2R_DroMel_CAF1 17042604 105 + 20766785 UUAGCAUGCGAUUUUGCUCGG----UUCGUG------AUUUCACAAAGCGGUGCGAAUCGA---CGGCCACAAGGGCUCACCUUUGUACGCCAAA--CAUGAAUAAUGCUCAUGAAUCGA ..(((((.((((((.(((((.----((.(((------....))).)).))).)))))))).---.(((.(((((((....)))))))..)))...--........))))).......... ( -31.00) >DroSec_CAF1 135906 107 + 1 UUAGCAUUCGAUU-UGUUCGG----UUCGUG------AUUUCACACAGCGGUGCGAAUCGACGACGGCCACAAGGGCUCACCUUUGUACGCCAAA--CAUGAAUAAUGCUCAUGAAUCGA .......((((((-(((.(.(----((.(((------......)))))).).)))))))))(((.(((.(((((((....)))))))..)))...--(((((.......)))))..))). ( -34.70) >DroSim_CAF1 144153 107 + 1 CUAGCAUUCGAUU-UGUUCGG----UUCGUG------AUUUCACAAAGCGGUGCGAAUCGACGACGGCCACAAGGGCUCACCUUUGUACGCCAAA--CAUGAAUAAUGCUCAUGAAUCGA .......((((((-((((((.----((.(((------....))).)).))).)))))))))(((.(((.(((((((....)))))))..)))...--(((((.......)))))..))). ( -35.20) >DroEre_CAF1 140100 107 + 1 CAAGCAUUCAAUU----UCGG----CUCGUGAUUGUGAUUUCACAAUGCGGCGCGAAUCGA---CGGCCACAAGGGCUCACCUUUGUACGCCGAA--CAUGAAUAAUGCUCAUGAAUCGA ............(----((((----(.(((.((((((....))))))))))).))))((((---((((.(((((((....)))))))..))))..--(((((.......)))))..)))) ( -36.40) >DroYak_CAF1 149597 101 + 1 UUGGCCAGCGAUU----UCGG----UUCGUG------AUUUCACAAUGCGGCGCGAAUCGA---CGGCCACAAGGGCUCACCUUUGUGCGCCAAA--CAUGAAUAAUGCUCAUGAAUCGA ........(((((----((((----((((((------.(..(.....)..)))))))))))---.(((((((((((....)))))))).)))...--(((((.......)))))))))). ( -36.30) >DroAna_CAF1 155494 106 + 1 AAAAAAAUAAAUA----CUGAACCAAUCAUG----ACAUUUCACAAGUUUGCGCAAAUUUU---UGGGAACAAUAGCCCACCAUUGUACGCCAAAAAGAUCAAUAAUAA---UGUCACAA .............----............((----(((((........(((......((((---((((.(((((........))))).).)))))))...)))....))---)))))... ( -14.30) >consensus UUAGCAUUCGAUU____UCGG____UUCGUG______AUUUCACAAAGCGGCGCGAAUCGA___CGGCCACAAGGGCUCACCUUUGUACGCCAAA__CAUGAAUAAUGCUCAUGAAUCGA ...((...(((......)))........(((..........)))...))........((((....(((.(((((((....)))))))..))).....(((((.......)))))..)))) (-13.17 = -13.40 + 0.23)
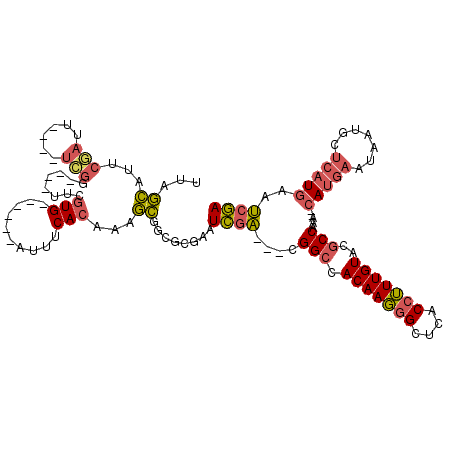
| Location | 17,042,604 – 17,042,709 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -18.12 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17042604 105 - 20766785 UCGAUUCAUGAGCAUUAUUCAUG--UUUGGCGUACAAAGGUGAGCCCUUGUGGCCG---UCGAUUCGCACCGCUUUGUGAAAU------CACGAA----CCGAGCAAAAUCGCAUGCUAA (((((.((((((.....))))))--...(((.(((((.((....)).)))))))))---))))...(((.((.((((((....------))))))----.)).((......)).)))... ( -30.40) >DroSec_CAF1 135906 107 - 1 UCGAUUCAUGAGCAUUAUUCAUG--UUUGGCGUACAAAGGUGAGCCCUUGUGGCCGUCGUCGAUUCGCACCGCUGUGUGAAAU------CACGAA----CCGAACA-AAUCGAAUGCUAA ..........((((((.....((--((((((.(((((.((....)).)))))))).((((.(((((((((....))))).)))------))))).----..)))))-.....)))))).. ( -33.70) >DroSim_CAF1 144153 107 - 1 UCGAUUCAUGAGCAUUAUUCAUG--UUUGGCGUACAAAGGUGAGCCCUUGUGGCCGUCGUCGAUUCGCACCGCUUUGUGAAAU------CACGAA----CCGAACA-AAUCGAAUGCUAG ..........((((((.....((--((((((.(((((.((....)).)))))))).((((.((((((((......)))).)))------))))).----..)))))-.....)))))).. ( -31.10) >DroEre_CAF1 140100 107 - 1 UCGAUUCAUGAGCAUUAUUCAUG--UUCGGCGUACAAAGGUGAGCCCUUGUGGCCG---UCGAUUCGCGCCGCAUUGUGAAAUCACAAUCACGAG----CCGA----AAUUGAAUGCUUG .........((((((...(((..--((((((.....((((.....))))(((((((---......)).)))))((((((....)))))).....)----))))----)..))))))))). ( -35.10) >DroYak_CAF1 149597 101 - 1 UCGAUUCAUGAGCAUUAUUCAUG--UUUGGCGCACAAAGGUGAGCCCUUGUGGCCG---UCGAUUCGCGCCGCAUUGUGAAAU------CACGAA----CCGA----AAUCGCUGGCCAA (((((.((((((.....))))))--...(((.(((((.((....)).)))))))))---))))...(.(((((.(((((....------))))).----..(.----...))).)))).. ( -31.70) >DroAna_CAF1 155494 106 - 1 UUGUGACA---UUAUUAUUGAUCUUUUUGGCGUACAAUGGUGGGCUAUUGUUCCCA---AAAAUUUGCGCAAACUUGUGAAAUGU----CAUGAUUGGUUCAG----UAUUUAUUUUUUU (..(((((---((..........(((((((.(.(((((((....))))))).))))---))))....((((....)))).)))))----))..).........----............. ( -24.90) >consensus UCGAUUCAUGAGCAUUAUUCAUG__UUUGGCGUACAAAGGUGAGCCCUUGUGGCCG___UCGAUUCGCACCGCUUUGUGAAAU______CACGAA____CCGA____AAUCGAAUGCUAA ((((..((((((.....))))))....((((.(((((.((....)).)))))))))...))))((((((......))))))....................................... (-18.12 = -17.98 + -0.13)

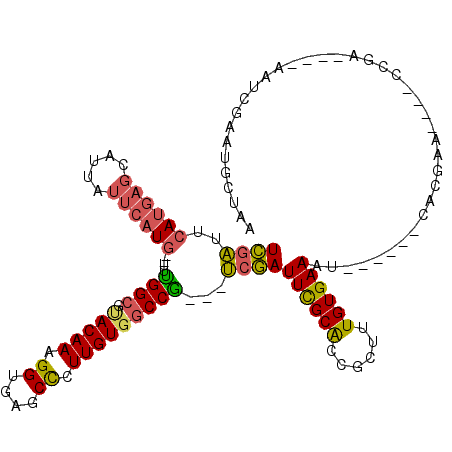
| Location | 17,042,634 – 17,042,749 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -28.50 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17042634 115 - 20766785 AAAGCCUUUGAAACAGAGGGCAGAACUGAACGAUUAUCGAUCGAUUCAUGAGCAUUAUUCAUGUUUGGCGUACAAAGGUGAGCCCUUGUGGCCG---UCGAUUCGCACCGCUUUGUGA ...((((((......)))))).........((.....))((((((.((((((.....))))))...(((.(((((.((....)).)))))))))---)))))(((((......))))) ( -31.60) >DroSec_CAF1 135935 116 - 1 AAAGCCUUUGAAACAGAGGGCAGAACUG--CGAUUAUCGAUCGAUUCAUGAGCAUUAUUCAUGUUUGGCGUACAAAGGUGAGCCCUUGUGGCCGUCGUCGAUUCGCACCGCUGUGUGA ....((((((...))))))((((...((--(((..(((((.((((.((((((.....))))))...(((.(((((.((....)).))))))))))))))))))))))...)))).... ( -40.80) >DroSim_CAF1 144182 116 - 1 AAAGCCUUUGAAACAGAGGGCAGAACUG--CGAUUAUCGAUCGAUUCAUGAGCAUUAUUCAUGUUUGGCGUACAAAGGUGAGCCCUUGUGGCCGUCGUCGAUUCGCACCGCUUUGUGA ...((((((......))))))((...((--(((..(((((.((((.((((((.....))))))...(((.(((((.((....)).))))))))))))))))))))))...))...... ( -38.30) >DroEre_CAF1 140132 112 - 1 AACACCUUUGAAACAGAGGGCA-AACUG--CGAUUAGCGAUCGAUUCAUGAGCAUUAUUCAUGUUCGGCGUACAAAGGUGAGCCCUUGUGGCCG---UCGAUUCGCGCCGCAUUGUGA ....((((((...))))))(((-(..((--((....(((((((((....((((((.....))))))(((.(((((.((....)).)))))))))---)))).))))..)))))))).. ( -40.10) >DroYak_CAF1 149623 112 - 1 AACACCUUUGAAACGGAGGGCA-CUCUG--CGAUUAGUGAUCGAUUCAUGAGCAUUAUUCAUGUUUGGCGCACAAAGGUGAGCCCUUGUGGCCG---UCGAUUCGCGCCGCAUUGUGA ....((((((...)))))).((-(..((--((....(((((((((.((((((.....))))))...(((.(((((.((....)).)))))))))---)))).))))..))))..))). ( -40.40) >consensus AAAGCCUUUGAAACAGAGGGCAGAACUG__CGAUUAUCGAUCGAUUCAUGAGCAUUAUUCAUGUUUGGCGUACAAAGGUGAGCCCUUGUGGCCG___UCGAUUCGCACCGCUUUGUGA ....((((((...))))))...................((((((..((((((.....))))))..((((.(((((.((....)).)))))))))...))))))((((......)))). (-28.50 = -27.78 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:46 2006