
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,019,546 – 17,019,756 |
| Length | 210 |
| Max. P | 0.993023 |

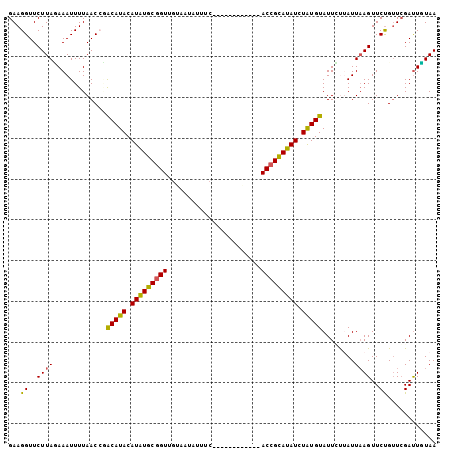
| Location | 17,019,546 – 17,019,650 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -13.89 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17019546 104 + 20766785 GAAGGUUCUUACAAAUUUUAACCAACAUACAUAUGCGGUUGUAAUAAUUCAAUUAUAA-CCGACCGCAUAUCUAUGUAUUGUUAUUAAGUUCUGUUCGAUUGUAA ......((..((((((((((((..(((((.(((((((((((..(((((...)))))..-.))))))))))).)))))...))))..))))).)))..))...... ( -24.10) >DroSec_CAF1 115753 92 + 1 GAAGGUUCUUAUAAAUUUUAACCGACAUACAUAUGCGGUUGUAAUAUUUC------------ACCGCAUAUCUGUGUAUUGUUAUU-AGUUCUGUUCGAUUGUAA ......((..(((((((.((((..(((((.(((((((((.(........)------------))))))))).)))))...))))..-)))).)))..))...... ( -19.80) >DroSim_CAF1 122591 93 + 1 GAAGGUUCUUAUAAAUUUUAACCGACAUACAUAUGCGGUUGUAAUAUUUC------------ACCGCAUAUCUGUGUAUUUUUAUUAAGUUCUGUUCGAUUGUAA (((((..((((((((.........(((((.(((((((((.(........)------------))))))))).)))))...)))).))))..)).)))........ ( -19.90) >DroEre_CAF1 118142 80 + 1 GAAGGUUCUUAGAAAUUUUAACAUACAUACAUAUGCAG-------------------------CCGUAUAUAUAUGUAUUCCAAUUAAGUUCUGAUGGACUCUAA ..((((((((((((........(((((((.((((((..-------------------------..)))))).)))))))..........)))))).)))).)).. ( -18.97) >DroYak_CAF1 124826 103 + 1 GAAGGUUCUUAGAAAUUUUAACACGCAUACAUGUGCGGUUGGGAUAU--GAGCUAUAUUCAGACCGUAUAUAUAUGUAUUCCAAUUAAGUUCCGUUCGAUUUUAA (((((..(((((............(((((.((((((((((.((((((--(...))))))).)))))))))).))))).......)))))..)).)))........ ( -26.41) >consensus GAAGGUUCUUAGAAAUUUUAACCGACAUACAUAUGCGGUUGUAAUAUUUC____________ACCGCAUAUCUAUGUAUUCUUAUUAAGUUCUGUUCGAUUGUAA ...((..((((.............(((((.(((((((((.......................))))))))).)))))........))))..))............ (-13.89 = -13.38 + -0.51)


| Location | 17,019,546 – 17,019,650 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -9.65 |
| Energy contribution | -10.74 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17019546 104 - 20766785 UUACAAUCGAACAGAACUUAAUAACAAUACAUAGAUAUGCGGUCGG-UUAUAAUUGAAUUAUUACAACCGCAUAUGUAUGUUGGUUAAAAUUUGUAAGAACCUUC ((((((((.....))......((((...(((((.(((((((((..(-(.(((((...))))).)).))))))))).)))))..))))....))))))........ ( -27.00) >DroSec_CAF1 115753 92 - 1 UUACAAUCGAACAGAACU-AAUAACAAUACACAGAUAUGCGGU------------GAAAUAUUACAACCGCAUAUGUAUGUCGGUUAAAAUUUAUAAGAACCUUC ..................-..((((...(((.(.(((((((((------------...........))))))))).).)))..)))).................. ( -17.80) >DroSim_CAF1 122591 93 - 1 UUACAAUCGAACAGAACUUAAUAAAAAUACACAGAUAUGCGGU------------GAAAUAUUACAACCGCAUAUGUAUGUCGGUUAAAAUUUAUAAGAACCUUC ....((((((................(((((....((((((((------------...........))))))))))))).))))))................... ( -16.53) >DroEre_CAF1 118142 80 - 1 UUAGAGUCCAUCAGAACUUAAUUGGAAUACAUAUAUAUACGG-------------------------CUGCAUAUGUAUGUAUGUUAAAAUUUCUAAGAACCUUC ((((((((((............))))((((((((((((....-------------------------....))))))))))))........))))))........ ( -16.20) >DroYak_CAF1 124826 103 - 1 UUAAAAUCGAACGGAACUUAAUUGGAAUACAUAUAUAUACGGUCUGAAUAUAGCUC--AUAUCCCAACCGCACAUGUAUGCGUGUUAAAAUUUCUAAGAACCUUC ........(((.((..(((((((..(((((.(((((((.((((..(.((((.....--)))).)..))))...))))))).)))))..)))...))))..))))) ( -16.40) >consensus UUACAAUCGAACAGAACUUAAUAACAAUACAUAGAUAUGCGGU____________GAAAUAUUACAACCGCAUAUGUAUGUCGGUUAAAAUUUAUAAGAACCUUC ............................(((((.(((((((((.......................))))))))).)))))........................ ( -9.65 = -10.74 + 1.09)

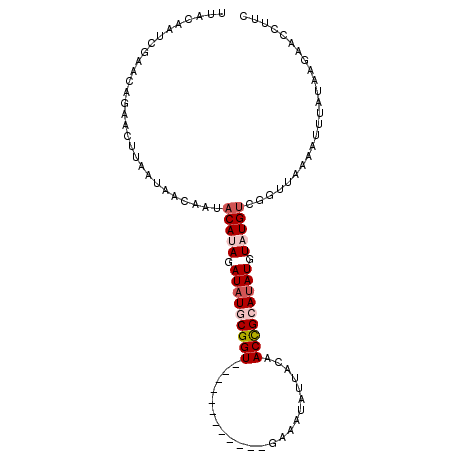

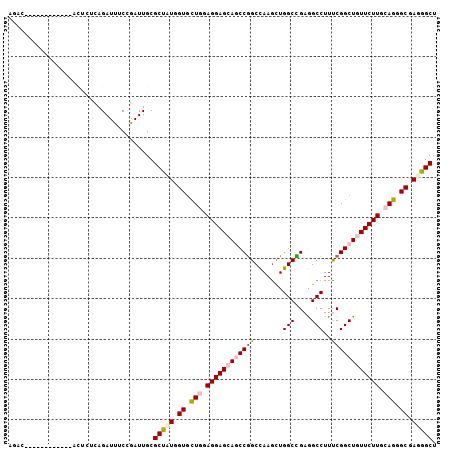
| Location | 17,019,650 – 17,019,756 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -33.89 |
| Energy contribution | -34.70 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17019650 106 + 20766785 UAAGCUUGUUUUUUCCACACACAGAUUUCCGAUUGCGCUUUGGUGCUGGAGGAGCAGCCGGCUAAGCUGGCCGAGGCCUUCCGGCUCUUCUUGCAGGGCGAGGGCU ....((((.(((((((((((.((((....((....)).))))))).))))))))..((((((...))))))))))((((((..(((((......))))))))))). ( -36.80) >DroVir_CAF1 170252 99 + 1 UGAAAUUUU-------UGCCACAGAUCUCGGAUUGCGCCAUGGUGCUGGAGGAGCAACCGGCUAAGCUGGCUGAGGCCUUUCGGCUGUUCUUGCAGGGCGAGGGCU .....((((-------((((..............((((....))))((.((((((..(((((...)))))....((((....)))))))))).)).)))))))).. ( -33.50) >DroPse_CAF1 132890 100 + 1 AUUC------CUUCUGCCUUUCAGAUUUCCGAUUGCGCUCUGGUGCUCGAGGAGCAGCCGGCCAAGCUGGCCGAGGCCUUUCGGCUGUUCUUGCAGGGCGAGGGCU ...(------((((.(((((.(.......(((..((((....)))))))((((((((((((((.....))))(((....)))))))))))))).)))))))))).. ( -44.00) >DroWil_CAF1 182358 90 + 1 ----------------AUUCUUAGAUUUCCGAUUGCGCUUUGGUUCUGGAGGAGCAGCCAGCCAAAUUGGCUGAAGCCUUUCGGCUAUUCUUGCAGGGCGAGGGCU ----------------....................(((((.((((((.((((((((((((.....))))))).((((....)))).))))).)))))).))))). ( -34.70) >DroMoj_CAF1 174669 99 + 1 AGAGUUUUU-------UUUUUUAGAUCUCUGAUUGCGCCAUGGUGUUGGAGGAGCAACCGGCUAAGCUGGCUGAAGCCUUCCGGCUGUUCUUGCAGGGCGAGGGCU ((((.(((.-------......))).))))......(((.(.((.(((.(((((((.((((....(((......)))...)))).))))))).))).)).).))). ( -34.30) >DroPer_CAF1 132138 100 + 1 AUUC------CUUCUGCCUUUCAGAUUUCCGAUUGCGCUCUGGUGCUCGAGGAGCAGCCGGCCAAGCUGGCCGAGGCCUUUCGGCUGUUCUUGCAGGGCGAGGGCU ...(------((((.(((((.(.......(((..((((....)))))))((((((((((((((.....))))(((....)))))))))))))).)))))))))).. ( -44.00) >consensus AGAC____________ACUCUCAGAUUUCCGAUUGCGCUAUGGUGCUGGAGGAGCAGCCGGCCAAGCUGGCCGAGGCCUUUCGGCUGUUCUUGCAGGGCGAGGGCU ....................................(((((.((.(((.((((((((((((.......(((....)))..)))))))))))).))).)).))))). (-33.89 = -34.70 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:37 2006