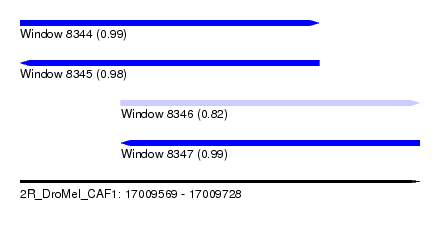
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,009,569 – 17,009,728 |
| Length | 159 |
| Max. P | 0.990784 |

| Location | 17,009,569 – 17,009,688 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.06 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17009569 119 + 20766785 UUUGAUAAGCGACGAUGUAUCGCUGGUCGGCGAUACUAUUUCUAAAAUAUCGGCUAGC-UCGAUAGCACCUAUAUGAAAACAUCGAUUCCCGAUGCAUCGAUGUUUCUCCCCAUCACUAU ...((.(((((.((((((((((..(((((((((((............)))))((((..-....))))...............))))))..)))))))))).))))).))........... ( -31.80) >DroSec_CAF1 105831 118 + 1 UUUGAAAAGCAACGAUGUAUCGCUGGGCUGCGAUACUAUUUCUAAAAUAUCGGCUAGCCUCGAUAGCAUCU--UUGAAAACAUCGAUUUCCGAUGCAUCGAUGUUUCUGCCCAUCACUAU .............((((((((((......)))))))...............(((.(((.(((((.(((((.--..((((.......)))).)))))))))).)))...))).)))..... ( -31.30) >DroSim_CAF1 112666 118 + 1 UUUGAAAAAUAACGAUGUAUCGCUGGGCUGCGAUACUAUUUCUAAAAUAUCGGCUAGCCUCGAUACCAUCU--UUGAAAACAUCGAUUCCCGAUGCAUCGAUGUUUCCGCCCAUCACUAU ..(((........(((((((((..(((((((((((............)))).)).)))))))))).)))).--..(.((((((((((.(.....).)))))))))).).....))).... ( -29.70) >DroEre_CAF1 107996 115 + 1 UUUGAAAAUCAACGAUGUAUCGCUGGUCGGCGAUA---UUACAGAAAUAUCGGCUAACAUCGAUAACAUCU--UUGAAAACAUCGAUUCUCGAUGCAUCGAUGUUUUUUCACAUCACUAU ..(((((.((((.(((((((((.((...(.(((((---((.....))))))).)...)).)))).))))).--))))((((((((((.(.....).)))))))))))))))......... ( -35.50) >DroYak_CAF1 114714 115 + 1 UUUUAAAAUCUACGAUGUAUCGUUGGUCAGCGAUA---UUACAGAAAUAACGGCCAACAUCGAUAGCAGCU--UUCAGAACAUCGAUUCUCGAUGCAUCGAUGUGCCUUCACAUCACUAG .............((((((((((((((...((.((---((.....)))).))))))))((((((..(....--....)...))))))....))))))))((((((....))))))..... ( -30.90) >consensus UUUGAAAAGCAACGAUGUAUCGCUGGUCGGCGAUACUAUUUCUAAAAUAUCGGCUAGCAUCGAUAGCAUCU__UUGAAAACAUCGAUUCCCGAUGCAUCGAUGUUUCUGCCCAUCACUAU .............((((((((((......))))))............((((((......))))))............((((((((((.(.....).)))))))))).....))))..... (-23.30 = -23.06 + -0.24)

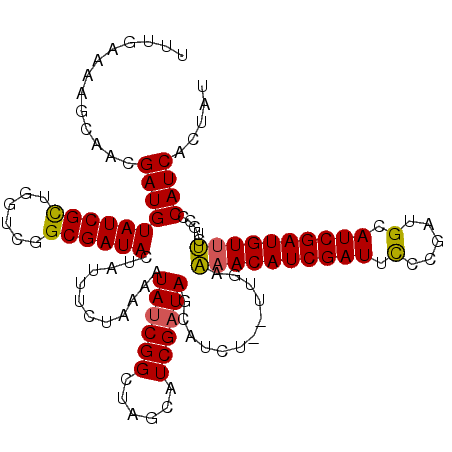
| Location | 17,009,569 – 17,009,688 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -24.21 |
| Energy contribution | -25.25 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17009569 119 - 20766785 AUAGUGAUGGGGAGAAACAUCGAUGCAUCGGGAAUCGAUGUUUUCAUAUAGGUGCUAUCGA-GCUAGCCGAUAUUUUAGAAAUAGUAUCGCCGACCAGCGAUACAUCGUCGCUUAUCAAA ..((((((((.(((((.(((((((.........))))))))))))((((.((((((....)-))..))).))))..........(((((((......))))))).))))))))....... ( -35.50) >DroSec_CAF1 105831 118 - 1 AUAGUGAUGGGCAGAAACAUCGAUGCAUCGGAAAUCGAUGUUUUCAA--AGAUGCUAUCGAGGCUAGCCGAUAUUUUAGAAAUAGUAUCGCAGCCCAGCGAUACAUCGUUGCUUUUCAAA ....(((..(((((((((((((((.........)))))))))))...--.((((.(((((.((((.((.((((((........))))))))))))...)))))))))..))))..))).. ( -42.10) >DroSim_CAF1 112666 118 - 1 AUAGUGAUGGGCGGAAACAUCGAUGCAUCGGGAAUCGAUGUUUUCAA--AGAUGGUAUCGAGGCUAGCCGAUAUUUUAGAAAUAGUAUCGCAGCCCAGCGAUACAUCGUUAUUUUUCAAA ..((((((((..((((((((((((.........))))))))))))..--.....((((((.((((.((.((((((........))))))))))))...)))))).))))))))....... ( -40.30) >DroEre_CAF1 107996 115 - 1 AUAGUGAUGUGAAAAAACAUCGAUGCAUCGAGAAUCGAUGUUUUCAA--AGAUGUUAUCGAUGUUAGCCGAUAUUUCUGUAA---UAUCGCCGACCAGCGAUACAUCGUUGAUUUUCAAA .........(((((((((((((((.........))))))))).((((--.(((((.((((..((..((.((((((.....))---))))))..))...))))))))).)))))))))).. ( -34.90) >DroYak_CAF1 114714 115 - 1 CUAGUGAUGUGAAGGCACAUCGAUGCAUCGAGAAUCGAUGUUCUGAA--AGCUGCUAUCGAUGUUGGCCGUUAUUUCUGUAA---UAUCGCUGACCAACGAUACAUCGUAGAUUUUAAAA .....((((((....))))))((.((((((.....))))))))((((--(.(((((((((..((..((.(((((....))))---)...))..))...)))))....)))).)))))... ( -30.70) >consensus AUAGUGAUGGGAAGAAACAUCGAUGCAUCGGGAAUCGAUGUUUUCAA__AGAUGCUAUCGAGGCUAGCCGAUAUUUUAGAAAUAGUAUCGCAGACCAGCGAUACAUCGUUGAUUUUCAAA ..((((((.....(((((((((((.........)))))))))))......((((.(((((........(((((((........)))))))........)))))))))))))))....... (-24.21 = -25.25 + 1.04)

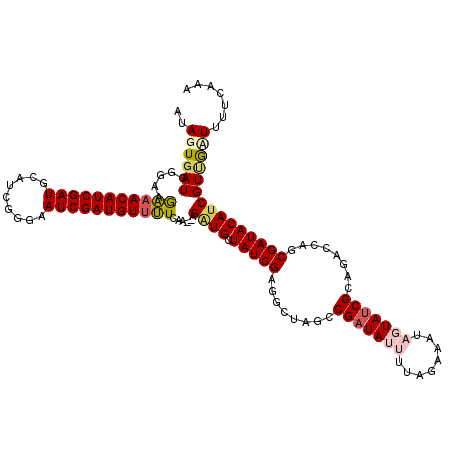
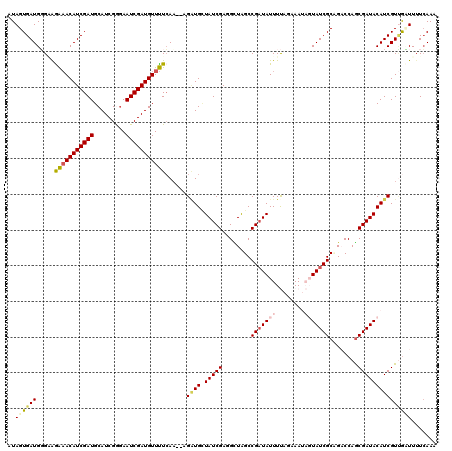
| Location | 17,009,609 – 17,009,728 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -20.84 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17009609 119 + 20766785 UCUAAAAUAUCGGCUAGC-UCGAUAGCACCUAUAUGAAAACAUCGAUUCCCGAUGCAUCGAUGUUUCUCCCCAUCACUAUCAAAUUUGCUUUGCGCUGCAUCGAGACCGAGAAUUAAAGA (((..(((.((((....(-(((((.(((.......((((((((((((.(.....).)))))))))).))..................((.....))))))))))).))))..)))..))) ( -26.60) >DroSec_CAF1 105871 118 + 1 UCUAAAAUAUCGGCUAGCCUCGAUAGCAUCU--UUGAAAACAUCGAUUUCCGAUGCAUCGAUGUUUCUGCCCAUCACUAUCAAAUUUGCUUUGCGCUGCAUCGAGACCGAGAAUUAAAGA (((..(((.((((((.((..((..((((..(--((((...(((((.....)))))....((((........))))....)))))..))))...))..))....)).))))..)))..))) ( -27.30) >DroSim_CAF1 112706 118 + 1 UCUAAAAUAUCGGCUAGCCUCGAUACCAUCU--UUGAAAACAUCGAUUCCCGAUGCAUCGAUGUUUCCGCCCAUCACUAUCAAAUUUGCUUUGCGCUGCAUCGAGACCGAGAAUUAAAGA .......((((((......))))))...(((--((((.....(((..((.(((((((..((((........))))............((.....))))))))).)).)))...))))))) ( -25.00) >DroEre_CAF1 108033 118 + 1 ACAGAAAUAUCGGCUAACAUCGAUAACAUCU--UUGAAAACAUCGAUUCUCGAUGCAUCGAUGUUUUUUCACAUCACUAUCAAAUUUGCUUUACGCUGCAUCGAGACCGAGAAUUAAAGA .......((((((......))))))...(((--((((.....(((..((((((((((..(((((......)))))............((.....)))))))))))).)))...))))))) ( -31.50) >DroYak_CAF1 114751 118 + 1 ACAGAAAUAACGGCCAACAUCGAUAGCAGCU--UUCAGAACAUCGAUUCUCGAUGCAUCGAUGUGCCUUCACAUCACUAGAAAAUUUGCUUUGCGCUGCAUCGAGACCGAGAAUUAAAGA ..........(((......(((((.((((((--(((.((.(((((.....)))))..))((((((....))))))....))))....((...))))))))))))..)))........... ( -31.90) >consensus UCUAAAAUAUCGGCUAGCAUCGAUAGCAUCU__UUGAAAACAUCGAUUCCCGAUGCAUCGAUGUUUCUGCCCAUCACUAUCAAAUUUGCUUUGCGCUGCAUCGAGACCGAGAAUUAAAGA .........((((......(((((....((.....))....))))).((.(((((((..((((........))))............((.....))))))))).)))))).......... (-20.84 = -21.48 + 0.64)

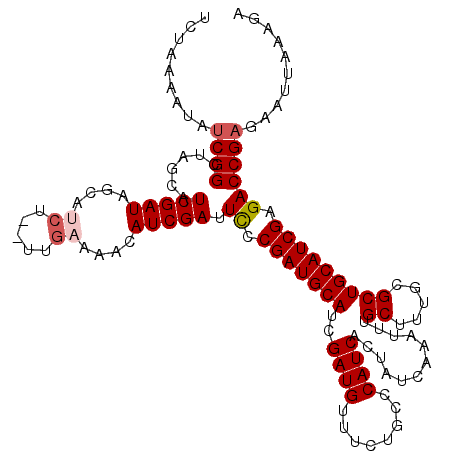
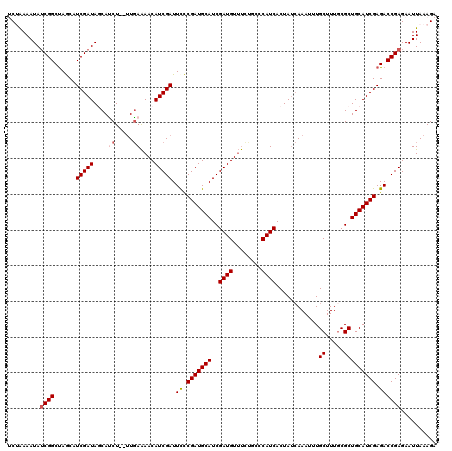
| Location | 17,009,609 – 17,009,728 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -35.96 |
| Consensus MFE | -29.22 |
| Energy contribution | -29.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17009609 119 - 20766785 UCUUUAAUUCUCGGUCUCGAUGCAGCGCAAAGCAAAUUUGAUAGUGAUGGGGAGAAACAUCGAUGCAUCGGGAAUCGAUGUUUUCAUAUAGGUGCUAUCGA-GCUAGCCGAUAUUUUAGA (((..(((..((((((((((((((.(.((((.....))))...........(((((.(((((((.........)))))))))))).....).))).)))))-)...))))).)))..))) ( -33.60) >DroSec_CAF1 105871 118 - 1 UCUUUAAUUCUCGGUCUCGAUGCAGCGCAAAGCAAAUUUGAUAGUGAUGGGCAGAAACAUCGAUGCAUCGGAAAUCGAUGUUUUCAA--AGAUGCUAUCGAGGCUAGCCGAUAUUUUAGA (((..(((..((((((((((((((((.((..((..........))..)).)).(((((((((((.........)))))))))))...--...))).))))))....))))).)))..))) ( -35.00) >DroSim_CAF1 112706 118 - 1 UCUUUAAUUCUCGGUCUCGAUGCAGCGCAAAGCAAAUUUGAUAGUGAUGGGCGGAAACAUCGAUGCAUCGGGAAUCGAUGUUUUCAA--AGAUGGUAUCGAGGCUAGCCGAUAUUUUAGA (((..(((..(((((((((((((.((.((..((..........))..)).))((((((((((((.........))))))))))))..--.....))))))))....))))).)))..))) ( -39.40) >DroEre_CAF1 108033 118 - 1 UCUUUAAUUCUCGGUCUCGAUGCAGCGUAAAGCAAAUUUGAUAGUGAUGUGAAAAAACAUCGAUGCAUCGAGAAUCGAUGUUUUCAA--AGAUGUUAUCGAUGUUAGCCGAUAUUUCUGU ..........((((((((((((((((.....))...........((((((......)))))).))))))))))(((((((...((..--.))...))))))).....))))......... ( -34.60) >DroYak_CAF1 114751 118 - 1 UCUUUAAUUCUCGGUCUCGAUGCAGCGCAAAGCAAAUUUUCUAGUGAUGUGAAGGCACAUCGAUGCAUCGAGAAUCGAUGUUCUGAA--AGCUGCUAUCGAUGUUGGCCGUUAUUUCUGU ...........(((((((((((((((((...))....((((....((((((....))))))((.((((((.....)))))))).)))--)))))).)))))....))))).......... ( -37.20) >consensus UCUUUAAUUCUCGGUCUCGAUGCAGCGCAAAGCAAAUUUGAUAGUGAUGGGAAGAAACAUCGAUGCAUCGGGAAUCGAUGUUUUCAA__AGAUGCUAUCGAGGCUAGCCGAUAUUUUAGA ..........((((((((((((((((.....))...........(((((........))))).)))))))))..((((((...((.....))...)))))).....)))))......... (-29.22 = -29.66 + 0.44)

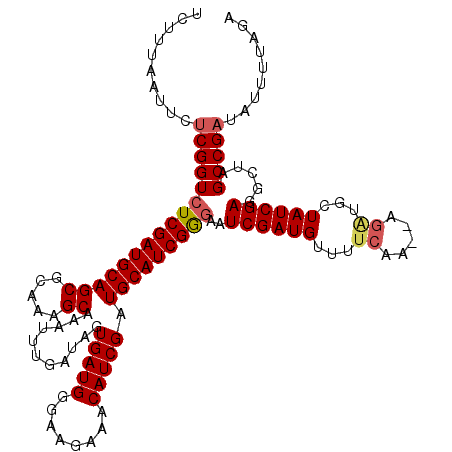
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:34 2006