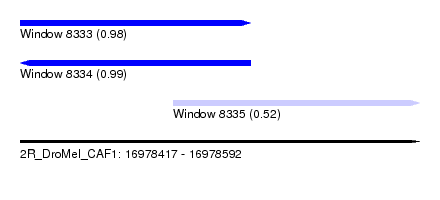
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,978,417 – 16,978,592 |
| Length | 175 |
| Max. P | 0.993628 |

| Location | 16,978,417 – 16,978,518 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16978417 101 + 20766785 ACCAGCUACCCAACCAUUCGCCACCCACUUACUCCCACAAACUCCUCUCGUCCACGCAACCAGCGUAAAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAA .((((..............(((..................((.......))..((((.....))))..............)))(....).))))....... ( -13.10) >DroSec_CAF1 75186 101 + 1 ACCAGCCUCCCAACCAUUCGCUACCCACUUACUACCAACAACUCCUCUUGUCCACGCAACCAGCGUAAAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAA ....(((............(((...............((((......))))..((((.....)))).........)))..((.(....).)))))...... ( -14.80) >DroSim_CAF1 80360 101 + 1 ACCAGCCACCCAACCAUUUGCCACCCACUUACUACCAACAACUCCUCUUGUCCACGCAACCAGCGUAAAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAA ....((((.........(((((.((..(((.......((((......))))..((((.....))))........)))...)).)))))...))))...... ( -17.90) >DroEre_CAF1 76546 101 + 1 ACCAGCCACCCAACCAUUCGCCACCCACUUACUACCAACAACUCCUCUUGUCCACGCAACCAGCGUAAAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAA ....((((...........(((...............((((......))))..((((.....))))..............)))(....)..))))...... ( -17.30) >DroYak_CAF1 83390 101 + 1 AAAAGCCAGCCAACCAUUCGCCACCCACUUACUACCAACAACUCCUCUUGUCCACGCAACCAGCGUAAAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAA ....(((((........(((((...............((((......))))..((((.....))))..............))))).....)))))...... ( -20.02) >consensus ACCAGCCACCCAACCAUUCGCCACCCACUUACUACCAACAACUCCUCUUGUCCACGCAACCAGCGUAAAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAA ....(((............(((...............((((......))))..((((.....))))..............)))(....)...)))...... (-14.22 = -14.50 + 0.28)

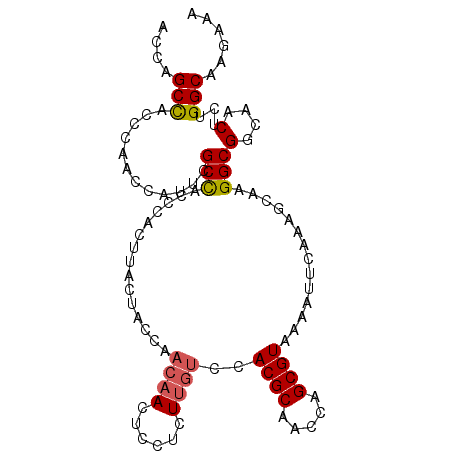
| Location | 16,978,417 – 16,978,518 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -29.86 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16978417 101 - 20766785 UUUCUUGCCAGAGUUGCCGCCUUGCUUUGAAUUUUACGCUGGUUGCGUGGACGAGAGGAGUUUGUGGGAGUAAGUGGGUGGCGAAUGGUUGGGUAGCUGGU ..(((.((((...(((((((((((((((.(..(((((((.....)))))))(....).......).))))))...))))))))).)))).)))........ ( -34.20) >DroSec_CAF1 75186 101 - 1 UUUCUUGCCAGAGUUGCCGCCUUGCUUUGAAUUUUACGCUGGUUGCGUGGACAAGAGGAGUUGUUGGUAGUAAGUGGGUAGCGAAUGGUUGGGAGGCUGGU (((((.((((..((((((..((((((........(((((.....)))))(((((......)))))...))))))..))))))...)))).)))))...... ( -32.20) >DroSim_CAF1 80360 101 - 1 UUUCUUGCCAGAGUUGCCGCCUUGCUUUGAAUUUUACGCUGGUUGCGUGGACAAGAGGAGUUGUUGGUAGUAAGUGGGUGGCAAAUGGUUGGGUGGCUGGU ..(((.((((...(((((((((.(((((....(((((((.....))))))).....)))))..............))))))))).)))).)))........ ( -31.66) >DroEre_CAF1 76546 101 - 1 UUUCUUGCCAGAGUUGCCGCCUUGCUUUGAAUUUUACGCUGGUUGCGUGGACAAGAGGAGUUGUUGGUAGUAAGUGGGUGGCGAAUGGUUGGGUGGCUGGU ..(((.((((...(((((((((.(((((....(((((((.....))))))).....)))))..............))))))))).)))).)))........ ( -31.36) >DroYak_CAF1 83390 101 - 1 UUUCUUGCCAGAGUUGCCGCCUUGCUUUGAAUUUUACGCUGGUUGCGUGGACAAGAGGAGUUGUUGGUAGUAAGUGGGUGGCGAAUGGUUGGCUGGCUUUU ...((.(((((..(((((((((.(((((....(((((((.....))))))).....)))))..............)))))))))....))))).))..... ( -31.86) >consensus UUUCUUGCCAGAGUUGCCGCCUUGCUUUGAAUUUUACGCUGGUUGCGUGGACAAGAGGAGUUGUUGGUAGUAAGUGGGUGGCGAAUGGUUGGGUGGCUGGU ...((.((((...(((((((((.(((((....(((((((.....))))))).....)))))..............))))))))).)))).))......... (-29.86 = -29.90 + 0.04)

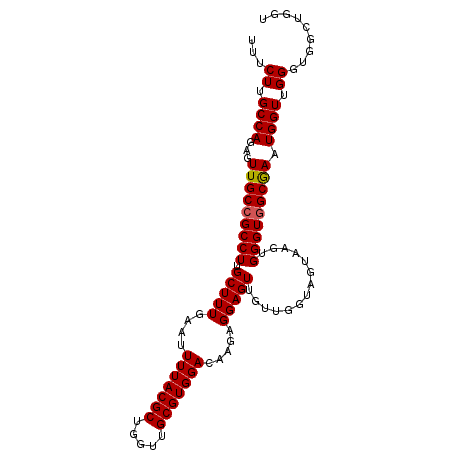
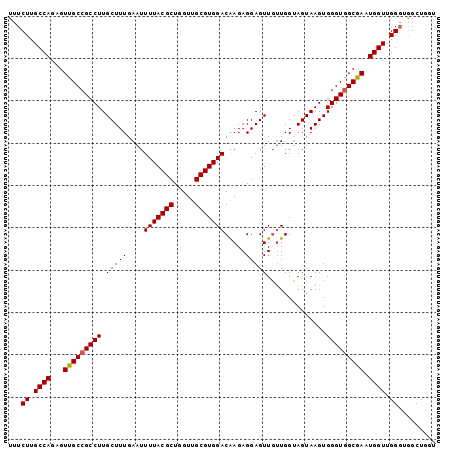
| Location | 16,978,484 – 16,978,592 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -15.44 |
| Energy contribution | -16.72 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16978484 108 + 20766785 AAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAAAAGAACAAAAUCUUGCUCCUAUUUCGGGCGUAAAUGAAAAUGCAAAUUUACUUUGAGCCGGGUGUCGCCAGCGG---- .........((..(((((((.(.(((((......((((......))))((.((......))))((((((..........)))))).....))))))))))))).))..---- ( -30.60) >DroPse_CAF1 85836 107 + 1 AAAUUCAAAGAAAAACGGAAA-AAUGGCAAGGGAAA---AAAAAUCUUGCUUCCAUUUGAGGCGUAAAUGAAAAUGUAAAUUCGCUUA-AAGCGCGUGCCACGAAUCAGACC ................(((..-...(((((((....---.....)))))))))).(((((..(((..((....))(((....(((...-..)))..))).)))..))))).. ( -19.50) >DroSim_CAF1 80427 107 + 1 AAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAAAAGAACAAAAUCUUGCUCCCAUUUCGGGCGUAAAUGAAAAUGCAAAUUCACUUU-AGCCGGGUGUCGCCAGCGG---- .........((..(((((((.(.(((((..................((((.(((.....))).)))).((((........))))....-.))))))))))))).))..---- ( -31.20) >DroEre_CAF1 76613 107 + 1 AAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAAAGGAACAAAAUCUUGCUCCCAUUUGAGGCGUAAAUGAAAAUGCAAAUUCACUUU-AGCCUGGUGUUGCCAGCGG---- ..............((((((((.(((((((((............)))))))........((((.((((((((........)))).)))-))))))).)))))).))..---- ( -28.40) >DroYak_CAF1 83457 107 + 1 AAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAAAAGAACAAAAUCUUGCUCCCAUUUGAGGCGUAAAUGAAAAUGCAAAUUCACUUU-AGCCGGGUGACGCCAGCGG---- .........((..((((...((((.(((((((............))))))).........(((.((((((((........)))).)))-))))))))..)))).))..---- ( -27.70) >DroPer_CAF1 88011 105 + 1 AAAUUCAAAGAAAAACGGAAA-AAUGGCAAGGGAAA-----AAAUCUUGCUUCCAUUUCAGGCGUAAAUGAAAAUGUUAAUUCGCUUA-AAGCGCGUGCCACGAAUCAGACC ..((((...((((...(((..-...(((((((....-----...)))))))))).)))).(((....((....))....((.(((...-..))).)))))..))))...... ( -19.50) >consensus AAAUUCAAAGCAAGGCGGCAACUCUGGCAAGAAAAAGAACAAAAUCUUGCUCCCAUUUCAGGCGUAAAUGAAAAUGCAAAUUCACUUU_AGCCGGGUGUCGCCAGCGG____ .............(((((((.....(((((((............))))))).........(((..((.((((........)))).))...)))...)))))))......... (-15.44 = -16.72 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:23 2006