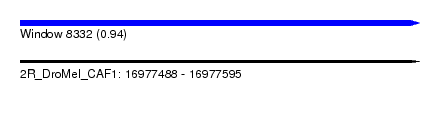
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,977,488 – 16,977,595 |
| Length | 107 |
| Max. P | 0.938985 |

| Location | 16,977,488 – 16,977,595 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.82 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -10.97 |
| Energy contribution | -12.45 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16977488 107 + 20766785 UAUCUAUCCACUGCCACU---GAGUUAGUUGAUUAAUUUUUCUGAAAUCUCAAAUCAAUUUAGCCUCAAGUGCAGCCAACAGCAAACUGCUGAUGGUGUUCUUCCC---AUCU------ ..........((((.(((---(((..((((((((........(((....)))))))))))....))).)))))))(((.((((.....)))).)))..........---....------ ( -22.90) >DroSec_CAF1 74212 106 + 1 UAUCUAUCCACUGCCGCU---GAGUUAGUUGAUUAAUUU-UGAGAAAUCUCAAAUCAAUUUAGCUUCAAGUGCAGCCAACAGCAAACUGCUGAUGGUU--C-CCCCUUCUUUU------ ..........((((.(((---(((((((((((((.....-((((....)))))))))).)))))))..)))))))(((.((((.....)))).)))..--.-...........------ ( -27.30) >DroSim_CAF1 79421 107 + 1 UAUCUAUCCACUGCCACU---GAGUUAGUUGAUUAAUUU-UGAGAAAUCGCAAAUCAAUUUAGCUUCAAGUGCAGCCAACAGCAAACUGCUGAUGGUU--CUCCCCUUCUUCU------ ..........((((.(((---(((((((((((((.....-(((....)))..)))))).)))))))..)))))))(((.((((.....)))).)))..--.............------ ( -25.80) >DroEre_CAF1 75533 109 + 1 UAU----UAACUGGCGCU---GAGUUAGCUGAUUAAUUU-UGAGAAAUCUCAAAUCAAUUUAGGUUCAAGUGCAUUACACAGCCAUCUGCUGACGGUU--CUGUCCCUCUUUUGCCACU ...----....(((((..---((((((((.(((....((-((((....))))))........((((...(((.....)))))))))).))))))((..--....)).))...))))).. ( -26.60) >DroYak_CAF1 82398 109 + 1 UAU----UAACUGCCAUU---GAAUUAGAUGAUUAAUUU-UGAGAAAUCUGAAAUCAAUUUAGGUUAGAGUGCAUCAAACAGCAAACUACUGAUGGUU--UUGUCCCUCUUUUGCCACC ...----........(((---((.((((((..(((....-)))...))))))..)))))...(((.((((.(.((.((((..((......))...)))--).)).)))))...)))... ( -16.70) >DroAna_CAF1 92355 106 + 1 UCUGG---AGCUGAGACUGGGGGGUUAGUUGAUUAAUUU-UGAGAAAUCUCAAAUCAAUUUAACUCCAAGUGUGUCG---------CUUCAGUUGCUGUUUUAUAUAUAUUUUUUCUCC .((((---(((.((.((...((((((((((((((.....-((((....)))))))))).))))))))....)).)))---------))))))........................... ( -30.90) >consensus UAUCU__CAACUGCCACU___GAGUUAGUUGAUUAAUUU_UGAGAAAUCUCAAAUCAAUUUAGCUUCAAGUGCAGCCAACAGCAAACUGCUGAUGGUU__CUCCCCCUCUUUU______ ...........((.((((...(((((((((((((......((((....)))))))))).)))))))..)))))).....((((.....))))........................... (-10.97 = -12.45 + 1.48)

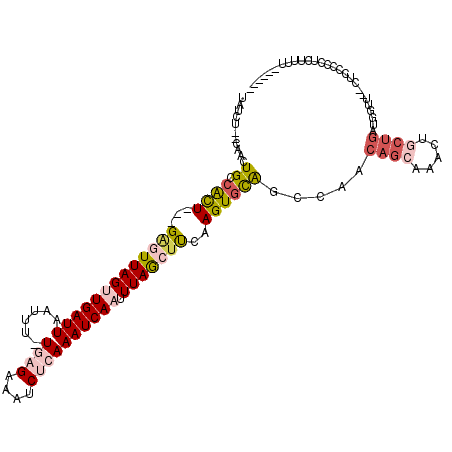

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:20 2006