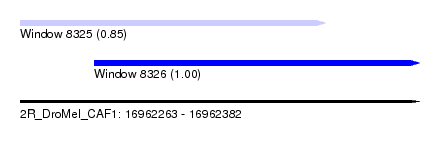
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,962,263 – 16,962,382 |
| Length | 119 |
| Max. P | 0.999822 |

| Location | 16,962,263 – 16,962,354 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -14.79 |
| Energy contribution | -14.97 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16962263 91 + 20766785 AAAUAUUUU--UUCGCGUGUGUGGUCCAUGUUUGCCAUCGGCAGACAUCGGAUAAUGUCUAAUAUAUUAUAUCCUAUUAACUGGAAAUACAAG .........--......(((((.(((((((((((((...))))))))).))))..................(((........))).))))).. ( -22.20) >DroSim_CAF1 63079 90 + 1 AAAUAUUUUUCUUAGCGUGUGUGGACCAUGUUUGCCAUCGGCAGACAUCGGAUGAUAUGUAAUAUAUUAUAUCCUAUUAACUGGAAAUAC--- .......(((((.((...(((.((((((((((((((...))))))))).))((((((((...)))))))).))))))...)))))))...--- ( -21.90) >DroEre_CAF1 60587 91 + 1 ACUUAU-UUUCUUCUAGUGUGUGAGCCAUAUUUGCCAUCGGCAGACAUUUGAUGAUGUGUAAUAUAUUGUAUCCUAUUAACUGGAAAUACCA- ......-....((((((((((.((((.(((((((((((((.(((....))).))))).)))).)))).)).)).)))..)))))))......- ( -17.00) >DroYak_CAF1 62636 93 + 1 CCUUGUGUUUUUUCUUGUGUAUGGGCCAUGUUUGCCAUCGGCAGACAUUUGAUUAUGUGUUUCAUAUUACAUCCUAUUAACUGGAAAUACAAG .(((((((((((....((..(((((..(((((((((...)))))))))......(((((........))))))))))..)).))))))))))) ( -25.90) >consensus AAAUAUUUUUCUUCGCGUGUGUGGGCCAUGUUUGCCAUCGGCAGACAUCGGAUGAUGUGUAAUAUAUUAUAUCCUAUUAACUGGAAAUACAA_ ..................((((...(((((((((((...))))))))....((((((((...))))))))...........)))..))))... (-14.79 = -14.97 + 0.19)

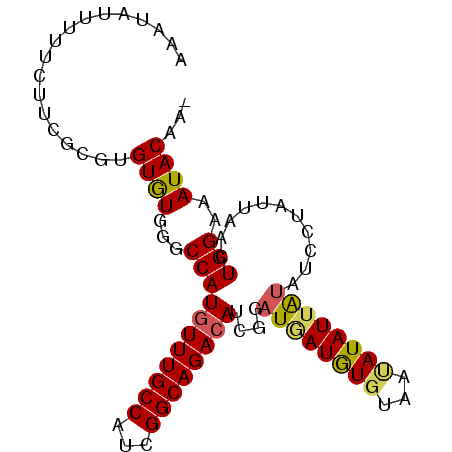
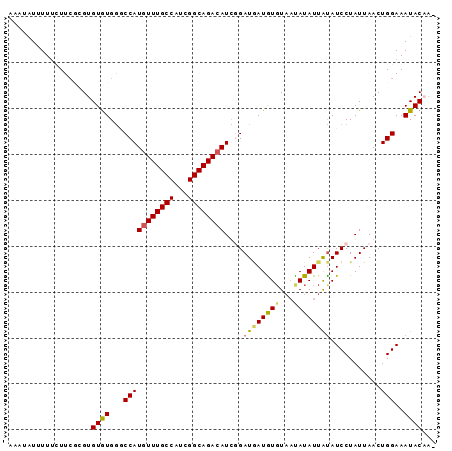
| Location | 16,962,285 – 16,962,382 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.23 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16962285 97 + 20766785 UCCAUGUUUGCCAUCGGCAGACAUCGGAUAAUGUCUAAUAUAUUAUAUCCUAUUAACUGGAAAUACAAGCUUCCAACUAAG-----------GCUAACAAGCCUAACA ((((((((((((...))))))))).)))((((((.....))))))............(((((........)))))....((-----------(((....))))).... ( -25.30) >DroSim_CAF1 63103 85 + 1 ACCAUGUUUGCCAUCGGCAGACAUCGGAUGAUAUGUAAUAUAUUAUAUCCUAUUAACUGGAAAUAC------------AAG-----------GCUAACAAGCCUAACA .(((((((((((...))))))))).))((((((((...)))))))).(((........))).....------------.((-----------(((....))))).... ( -24.90) >DroEre_CAF1 60610 76 + 1 GCCAUAUUUGCCAUCGGCAGACAUUUGAUGAUGUGUAAUAUAUUGUAUCCUAUUAACUGGAAAUACCA--------------------------------GCCCAACA ((.(((((((((((((.(((....))).))))).)))).)))).))..........((((.....)))--------------------------------)....... ( -13.30) >DroYak_CAF1 62660 108 + 1 GCCAUGUUUGCCAUCGGCAGACAUUUGAUUAUGUGUUUCAUAUUACAUCCUAUUAACUGGAAAUACAAGCUUUAUACUAACCCUAUGUAUCAUCUAAAAAGCCUAACA ...(((((((((...))))))))).......((((((((...(((........)))...)))))))).(((((((((.........)))).......)))))...... ( -19.91) >consensus GCCAUGUUUGCCAUCGGCAGACAUCGGAUGAUGUGUAAUAUAUUAUAUCCUAUUAACUGGAAAUACAA__________AAG___________GCUAACAAGCCUAACA ...(((((((((...)))))))))...((((((((...)))))))).(((........)))............................................... (-12.60 = -12.97 + 0.37)

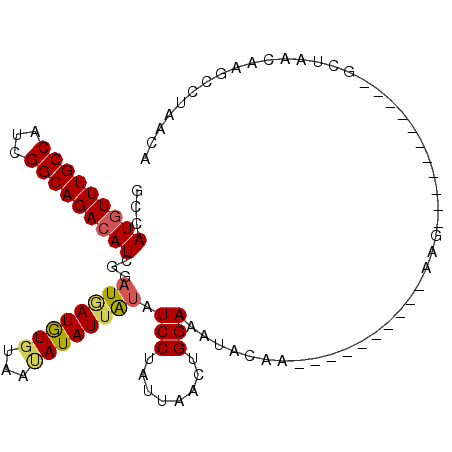
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:14 2006