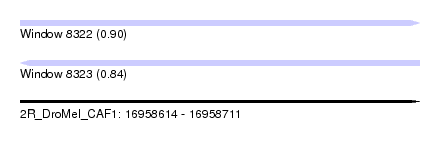
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,958,614 – 16,958,711 |
| Length | 97 |
| Max. P | 0.897435 |

| Location | 16,958,614 – 16,958,711 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -12.08 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
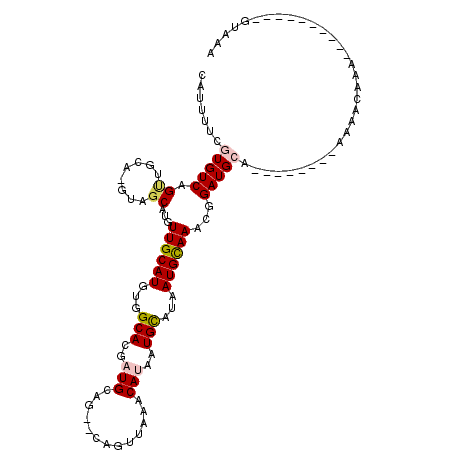
>2R_DroMel_CAF1 16958614 97 + 20766785 CAUUUUCGUGUCAGUUUCAUGUAGCAUAUUGCAUGUGGCACGAUGCAGUGCAGUUAAACAUAAUGCAUAAUGCAAACGGAUGCA--------AAAACAAA----------GUAAA (((((((((((((....(((((((....)))))))))))))))(((((((((...........)))))..))))...)))))..--------........----------..... ( -23.90) >DroPse_CAF1 65019 100 + 1 CAUUUGCAUGUCAGCUGUA-GUUGCACGUUGCAUGUGGCACGCUGCAC--CAGUGCCACAUAAUGCAUAAUGCAAGCGGAUGCAUU--GCCAGAAAUAAA----------GUAAA .....((((((...((((.-.(((((...((((((((((((.......--..))))))))...))))...)))))))))..))).)--))..........----------..... ( -30.90) >DroEre_CAF1 57039 97 + 1 CAUUUUCGUGUCAGUUACAUGUAGCAUGUUGCAUGUGGCACGAUGCAGUGCAGUUAAGCAUAAUGCAUAAUGCAAACGGAUGCA--------AAAACAAA----------GUAAA .......(((((.(((((((((((....)))))))).(((..(((((((((......))))..)))))..))).))).))))).--------........----------..... ( -28.10) >DroYak_CAF1 58161 92 + 1 UAUUUUCGUGUCAGUUUCAUGUAGCAUGUUGCAUGUGGCACGAUGC-----AGUUAAACAUAAUGCUUUAUGCAAACUGAUGCA--------AAAACAAA----------GUAAA .......((((((((((((((.(((((.((((((........))))-----)).........))))).)))).)))))))))).--------........----------..... ( -24.60) >DroAna_CAF1 60188 106 + 1 CAUUUUCGUGUCAG-------CGGCGAGUUGCAUGUGGCACGAUGCAG--CAGUUAAACAUAAUGUAUAAUGUAAGCGGAUGCACUACGCCAAAAACAAAACAAAAAAAGGAAAA ..(((((((....)-------)(((((((.((((.(((((..(((((.--.............)))))..)))...)).))))))).))))..................))))). ( -23.24) >DroPer_CAF1 67197 100 + 1 CAUUUGCAUGUCAGCUGUA-GUUGCACGUUGCAUGUGGCACGCUGCAC--CAGUGCCACAUAAUGCAUAAUGCAAGCGGAUGCAUU--GCCAGAAAUAAA----------GUAAA .....((((((...((((.-.(((((...((((((((((((.......--..))))))))...))))...)))))))))..))).)--))..........----------..... ( -30.90) >consensus CAUUUUCGUGUCAGUUGCA_GUAGCAUGUUGCAUGUGGCACGAUGCAG__CAGUUAAACAUAAUGCAUAAUGCAAACGGAUGCA________AAAACAAA__________GUAAA .......(((((.((........))...((((((...(((..(((.............)))..)))...))))))...)))))................................ (-12.08 = -12.47 + 0.39)


| Location | 16,958,614 – 16,958,711 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -12.57 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
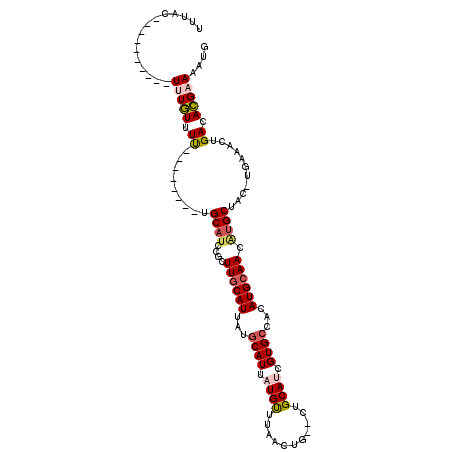
>2R_DroMel_CAF1 16958614 97 - 20766785 UUUAC----------UUUGUUUU--------UGCAUCCGUUUGCAUUAUGCAUUAUGUUUAACUGCACUGCAUCGUGCCACAUGCAAUAUGCUACAUGAAACUGACACGAAAAUG .....----------...(((((--------((..((.((((.(((...((((..(((......((((......)))).....)))..))))...))))))).))..))))))). ( -20.30) >DroPse_CAF1 65019 100 - 1 UUUAC----------UUUAUUUCUGGC--AAUGCAUCCGCUUGCAUUAUGCAUUAUGUGGCACUG--GUGCAGCGUGCCACAUGCAACGUGCAAC-UACAGCUGACAUGCAAAUG .....----------..(((.((.(((--((((((......)))))).(((((((((((((((..--.......))))))))))....)))))..-....))))).)))...... ( -28.70) >DroEre_CAF1 57039 97 - 1 UUUAC----------UUUGUUUU--------UGCAUCCGUUUGCAUUAUGCAUUAUGCUUAACUGCACUGCAUCGUGCCACAUGCAACAUGCUACAUGUAACUGACACGAAAAUG .....----------...(((((--------((..((.((((((((...((((.((((...........)))).))))...)))))(((((...)))))))).))..))))))). ( -21.90) >DroYak_CAF1 58161 92 - 1 UUUAC----------UUUGUUUU--------UGCAUCAGUUUGCAUAAAGCAUUAUGUUUAACU-----GCAUCGUGCCACAUGCAACAUGCUACAUGAAACUGACACGAAAAUA .....----------..((((((--------((..(((((((((((...((((.((((......-----)))).))))...))))..((((...)))))))))))..)))))))) ( -25.60) >DroAna_CAF1 60188 106 - 1 UUUUCCUUUUUUUGUUUUGUUUUUGGCGUAGUGCAUCCGCUUACAUUAUACAUUAUGUUUAACUG--CUGCAUCGUGCCACAUGCAACUCGCCG-------CUGACACGAAAAUG ........(((((((...(((..(((((.(((((((..((..((((........))))......)--).((.....))...)))).))))))))-------..)))))))))).. ( -22.00) >DroPer_CAF1 67197 100 - 1 UUUAC----------UUUAUUUCUGGC--AAUGCAUCCGCUUGCAUUAUGCAUUAUGUGGCACUG--GUGCAGCGUGCCACAUGCAACGUGCAAC-UACAGCUGACAUGCAAAUG .....----------..(((.((.(((--((((((......)))))).(((((((((((((((..--.......))))))))))....)))))..-....))))).)))...... ( -28.70) >consensus UUUAC__________UUUGUUUU________UGCAUCCGCUUGCAUUAUGCAUUAUGUUUAACUG__CUGCAUCGUGCCACAUGCAACAUGCUAC_UGAAACUGACACGAAAAUG ...............(((((.((.........((((....((((((...((((.((((...........)))).))))...)))))).))))...........)).))))).... (-12.57 = -12.93 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:11 2006