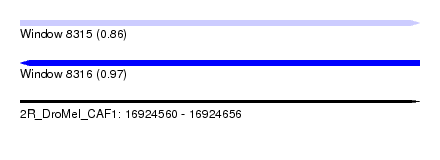
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,924,560 – 16,924,656 |
| Length | 96 |
| Max. P | 0.972678 |

| Location | 16,924,560 – 16,924,656 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16924560 96 + 20766785 AGAGACGUAGAUUAU-----GAAGGAGCGAGCCAAAGUGCUCGCUCGUGCCAAAAUAACGAACCGAACACUUCACCAGAAAGUUUCAAACUGACACCAAAA .(((((........(-----((((((((((((......))))))))((.........))..........))))).......)))))............... ( -24.56) >DroSec_CAF1 22157 96 + 1 AGAGACGUAGAUUAC-----GAAGGAGCGAGCCAAUGUGCUCGCUCGUGCCAAAAUAACCAACCGAACACUUCACCAGAAAGUUUCAAACUGACACCAAAA .((((((((...)))-----((((((((((((......))))))))((..................)).))))........)))))............... ( -23.37) >DroSim_CAF1 22213 96 + 1 AGAGACGUAGAUUAC-----GAAGGAGCGAGCCAAUGUGCUCGCUCGUGCCAAAAUAACCAACCGAACACUUCACCAGAAAGUUUCAAACUGACACCAAAA .((((((((...)))-----((((((((((((......))))))))((..................)).))))........)))))............... ( -23.37) >DroEre_CAF1 22561 96 + 1 AGAGACGUAGAUUAU-----GAAGGAGCGAGCCAAUGUGCUCGCUCGUGCCAAAAUAACCAACCGAACACUUCACCAGAAAGUUUCAAACUGACACCAAAA .(((((........(-----((((((((((((......))))))))((..................)).))))).......)))))............... ( -23.83) >DroYak_CAF1 22921 96 + 1 AGAGACGUAGAUUAU-----GAAGGAGCGAGCCACAGUGCUCGCUCGUGCCAAAAUAACCAACCGAACACUUCACCAGAAAGUUUCAAACUGACACCAAAA .(((((........(-----((((((((((((......))))))))((..................)).))))).......)))))............... ( -23.83) >DroAna_CAF1 24125 100 + 1 AGAAACGUAGAUAUUCAGGAGGAGGAGCGAGCCAGGACGCCCGCUCGAGCCAAAA-AAUCAACCGACCACUUCACCAGAAUGUUUUAAACUGACACCAAAA ........((((((((.((.(((((((((.((......)).)))))((.......-..)).........)))).)).))))))))................ ( -22.10) >consensus AGAGACGUAGAUUAU_____GAAGGAGCGAGCCAAUGUGCUCGCUCGUGCCAAAAUAACCAACCGAACACUUCACCAGAAAGUUUCAAACUGACACCAAAA .(((((..................((((((((......))))))))........................(((....))).)))))............... (-19.48 = -19.37 + -0.11)


| Location | 16,924,560 – 16,924,656 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.47 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16924560 96 - 20766785 UUUUGGUGUCAGUUUGAAACUUUCUGGUGAAGUGUUCGGUUCGUUAUUUUGGCACGAGCGAGCACUUUGGCUCGCUCCUUC-----AUAAUCUACGUCUCU .....(((((((..(((......((((........))))....)))..)))))))((((((((......))))))))....-----............... ( -27.80) >DroSec_CAF1 22157 96 - 1 UUUUGGUGUCAGUUUGAAACUUUCUGGUGAAGUGUUCGGUUGGUUAUUUUGGCACGAGCGAGCACAUUGGCUCGCUCCUUC-----GUAAUCUACGUCUCU .....(((((((.....((((..((((........))))..))))...)))))))((((((((......))))))))...(-----(((...))))..... ( -29.80) >DroSim_CAF1 22213 96 - 1 UUUUGGUGUCAGUUUGAAACUUUCUGGUGAAGUGUUCGGUUGGUUAUUUUGGCACGAGCGAGCACAUUGGCUCGCUCCUUC-----GUAAUCUACGUCUCU .....(((((((.....((((..((((........))))..))))...)))))))((((((((......))))))))...(-----(((...))))..... ( -29.80) >DroEre_CAF1 22561 96 - 1 UUUUGGUGUCAGUUUGAAACUUUCUGGUGAAGUGUUCGGUUGGUUAUUUUGGCACGAGCGAGCACAUUGGCUCGCUCCUUC-----AUAAUCUACGUCUCU .....(((((((.....((((..((((........))))..))))...)))))))((((((((......))))))))....-----............... ( -29.40) >DroYak_CAF1 22921 96 - 1 UUUUGGUGUCAGUUUGAAACUUUCUGGUGAAGUGUUCGGUUGGUUAUUUUGGCACGAGCGAGCACUGUGGCUCGCUCCUUC-----AUAAUCUACGUCUCU .....(((((((.....((((..((((........))))..))))...)))))))((((((((......))))))))....-----............... ( -29.40) >DroAna_CAF1 24125 100 - 1 UUUUGGUGUCAGUUUAAAACAUUCUGGUGAAGUGGUCGGUUGAUU-UUUUGGCUCGAGCGGGCGUCCUGGCUCGCUCCUCCUCCUGAAUAUCUACGUUUCU ....((((((((.............(((.(((..(((....))).-.))).))).((((((((......))))))))......))).)))))......... ( -25.10) >consensus UUUUGGUGUCAGUUUGAAACUUUCUGGUGAAGUGUUCGGUUGGUUAUUUUGGCACGAGCGAGCACAUUGGCUCGCUCCUUC_____AUAAUCUACGUCUCU .....(((((((.....((((..((((........))))..))))...)))))))((((((((......))))))))........................ (-25.94 = -26.47 + 0.53)

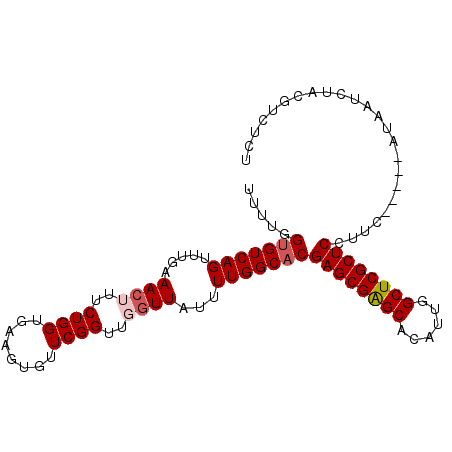
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:04 2006