
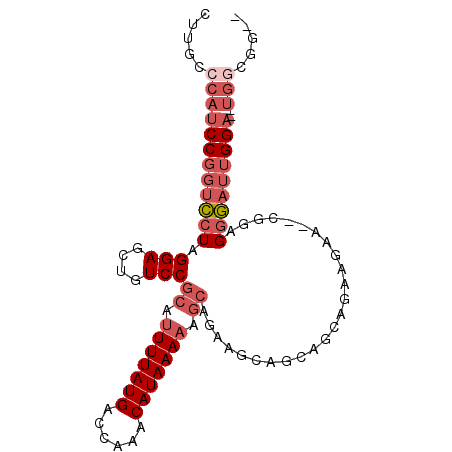
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,920,584 – 16,920,678 |
| Length | 94 |
| Max. P | 0.985293 |

| Location | 16,920,584 – 16,920,678 |
|---|---|
| Length | 94 |
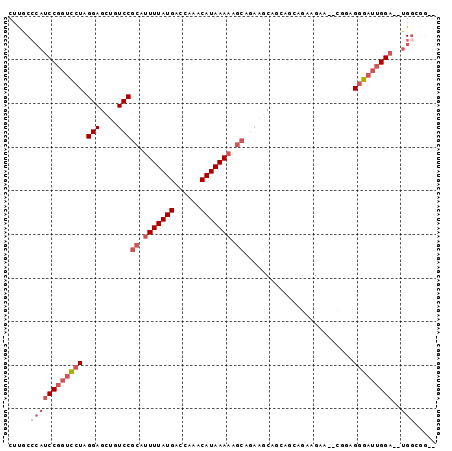
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -17.86 |
| Energy contribution | -19.58 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
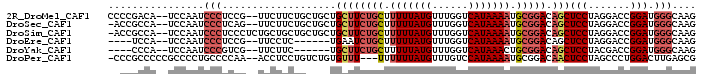
>2R_DroMel_CAF1 16920584 94 + 20766785 CUUGCCCAUCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGAAGAA--CGGAGGGAUUGGA--UGUCGGGG ((((..(((((((((((....((((((.((.(((((((......))))))).)).)..)))))....(.....--)...)))))))))--)).)))). ( -29.90) >DroSec_CAF1 18155 93 + 1 CUUGCCCAUCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGAAGAA--CUGAGGGAUUGGA--UGGCGGU- .(((((.((((((((((....((((((.((.(((((((......))))))).)).)..)))))..(((.....--))).)))))))))--)))))).- ( -35.40) >DroSim_CAF1 18199 95 + 1 CUUGCCCAUCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGCAGCAGAGGGAGGGAUUGGA--UGGCGGU- .(((((.((((((((((....((((((.((.(((((((......))))))).)).)..))))).((....)).......)))))))))--)))))).- ( -35.60) >DroEre_CAF1 18606 84 + 1 CUUGCCCAUCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAUUCA------GAGGAA--CGGAGGGAUUGGA--UGGA---- .....((((((((((((.(((....)))((.(((((((......))))))).)).......------......--....)))))))))--))).---- ( -30.00) >DroYak_CAF1 18950 84 + 1 CUUGCCCAUCCGGUCGUAGGAGCUGUCCGCAGUUUAUGACCAAACAUAAAAAGCAGAAGCA------GAAGAA--CGACGGGAUUGGA--UGGG---- ....(((((((((((((((..((((....))))))))))))...........((....)).------......--..........)))--))))---- ( -28.10) >DroPer_CAF1 24483 92 + 1 CGCUCAAGUCCAGGGCUAGGAGUUGUCCGCAUUUUAUGGACAAACAUAAAAAA---AAACACAGACAGGAGGU--UUGGGGCAGGGGCGGGGGCGGG- (((((..((((.((((........))))((.(((((((......)))))))..---.....(((((.....))--)))..))..))))..)))))..- ( -32.50) >consensus CUUGCCCAUCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGAAGAA__CGGAGGGAUUGGA__UGGCGG__ .....((((((((((((.(((....)))((.(((((((......))))))).)).........................)))))))))..)))..... (-17.86 = -19.58 + 1.72)

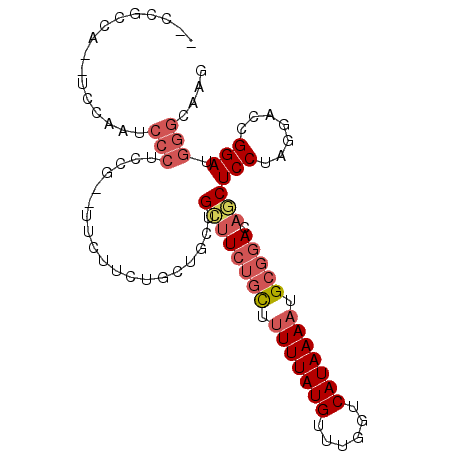
| Location | 16,920,584 – 16,920,678 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -14.53 |
| Energy contribution | -15.67 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16920584 94 - 20766785 CCCCGACA--UCCAAUCCCUCCG--UUCUUCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGAUGGGCAAG ........--......(((((((--.((((..(..((((..(((((.(((((((......))))))).)))))))))..).)))).)))).))).... ( -30.90) >DroSec_CAF1 18155 93 - 1 -ACCGCCA--UCCAAUCCCUCAG--UUCUUCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGAUGGGCAAG -...((((--(((..(((..(((--(......))))..((((((((.(((((((......))))))).))))).))).....)))..)))).)))... ( -31.70) >DroSim_CAF1 18199 95 - 1 -ACCGCCA--UCCAAUCCCUCCCUCUGCUGCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGAUGGGCAAG -...((((--(((..(((........((....)).((((..(((((.(((((((......))))))).))))))))).....)))..)))).)))... ( -31.80) >DroEre_CAF1 18606 84 - 1 ----UCCA--UCCAAUCCCUCCG--UUCCUC------UGAAUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGAUGGGCAAG ----....--......(((((((--.(((((------((..(((((.(((((((......))))))).)))))))).....)))).)))).))).... ( -28.10) >DroYak_CAF1 18950 84 - 1 ----CCCA--UCCAAUCCCGUCG--UUCUUC------UGCUUCUGCUUUUUAUGUUUGGUCAUAAACUGCGGACAGCUCCUACGACCGGAUGGGCAAG ----((((--(((......((((--(.....------.((((((((..((((((......))))))..))))).)))....))))).))))))).... ( -31.00) >DroPer_CAF1 24483 92 - 1 -CCCGCCCCCGCCCCUGCCCCAA--ACCUCCUGUCUGUGUUU---UUUUUUAUGUUUGUCCAUAAAAUGCGGACAACUCCUAGCCCUGGACUUGAGCG -..(((..((((.........((--(((........).))))---..(((((((......))))))).)))).(((.(((.......))).))).))) ( -16.80) >consensus __CCGCCA__UCCAAUCCCUCCG__UUCUUCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGAUGGGCAAG ................(((...................((((((((.(((((((......))))))).))))).)))(((.......))).))).... (-14.53 = -15.67 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:02 2006