
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,917,463 – 16,917,559 |
| Length | 96 |
| Max. P | 0.661609 |

| Location | 16,917,463 – 16,917,559 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -21.46 |
| Energy contribution | -20.97 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
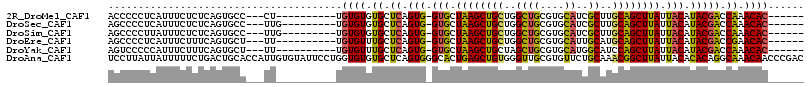
>2R_DroMel_CAF1 16917463 96 + 20766785 ACCCCCUCAUUUCUCUCAGUGCC---CU----------UGUGUGUGCUCAGUG-GUGCUAAGCUGCUGGCUGCGUGCAUCGCUUGCAGCUUAUUACAUACGACCAAACAC------ ..................((...---.(----------((((((((..(((..-((.....))..)))((((((.((...)).))))))....)))))))))....))..------ ( -27.60) >DroSec_CAF1 15118 97 + 1 AGCCCCUCAUUUCUCUCAGUGCC---UUG---------UGUGUGUGCUCAGUG-GUGCUAAGCUGCUGGCUGCGUGCAUCGCUUGCAGCUUAUUACAUACGACCAAACAC------ ..................((...---(((---------((((((.((((((((-((((..(((.....)))....))))))).)).)))....)))))))))....))..------ ( -27.70) >DroSim_CAF1 15123 97 + 1 AGCCCCUUAUUUCUCUCAGUGCC---UUG---------UGUGUGUGCUCAGUG-GUGCUAAGCUGCUGGCUGCGUGCAUCGCUUGCAGCUUAUUACAUACGACCAAACAC------ ..................((...---(((---------((((((.((((((((-((((..(((.....)))....))))))).)).)))....)))))))))....))..------ ( -27.70) >DroEre_CAF1 15459 96 + 1 AGCCCCUCAUUUCUUUCAGUGCU---UU----------UGUGUUUGCUCAGUG-GUGCUAAGCUGCUGGCUGCGUGCAUUGCAUGCAGCUUAUUACAUACGACCGAACAC------ .......................---..----------.(((((((.((((..-((.....))..))((((((((((...))))))))))..........)).)))))))------ ( -29.00) >DroYak_CAF1 15798 96 + 1 AGUCCCCCAUUUCUUUCAGUGCU---UU----------UGUGUUUGCUCAGUG-GUGCUAAGCUGCUAGCUGCGUGCAUGGCAUCCAGCUUAUUACAUACGACCAAACAC------ .......................---..----------.(((((((.((.(((-(((.(((((((...(((........)))...))))))).))).))))).)))))))------ ( -23.80) >DroAna_CAF1 16881 116 + 1 UCCUUAUUAUUUUUCUGACUGCACCAUUGUGUAUUCCUGGUGUGUGCUCAGUGGGCACUGAGCUGUGGGUUGCGUGUUCUGCAAACGGCUUAUUACACACAGGCAAACAACCCGAC ...................(((((....)))))..((((.((((((((((((....))))))).((((((((..((.....))..)))))))).)))))))))............. ( -32.40) >consensus AGCCCCUCAUUUCUCUCAGUGCC___UU__________UGUGUGUGCUCAGUG_GUGCUAAGCUGCUGGCUGCGUGCAUCGCAUGCAGCUUAUUACAUACGACCAAACAC______ .......................................((((.((.((.(((.(((.((((((((..((((....))..))..)))))))).))).))))).)).))))...... (-21.46 = -20.97 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:00 2006