
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,877,869 – 16,877,961 |
| Length | 92 |
| Max. P | 0.999860 |

| Location | 16,877,869 – 16,877,961 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16877869 92 + 20766785 ACUGCGGAGUGCCAGGACCACAGUGAAGACCGACCUCAAACUGGUCCUGUACCCCG------UCCGCCGUCGAUUUCCUCACUCCGUAGCAUCAAUCC .((((((((((.((((((((...(((.(......))))...)))))))).....((------.(....).)).......))))))))))......... ( -31.70) >DroSec_CAF1 28223 77 + 1 ACUGCGGAGUGCCAGGACCACAGUGAAGACCGACCACAAACUGGUCCUGUCCCCCACU---------------------CACUCCGGACCAUCAAUCC ....(((((((.((((((((..(((.........)))....)))))))).........---------------------)))))))............ ( -23.70) >DroSim_CAF1 28531 94 + 1 ACUGCGGAGUGCCAGGACCACAGUGAAGACCGACCUCAAACUGGUCCUGUCUCCCACU----CCCUCCGCCGAUUUGCUCACUCCGGACCAUCAAUCC ....(((((((...(((....((((.((((.((((.......))))..))))..))))----...)))((......)).)))))))............ ( -28.10) >DroEre_CAF1 28591 80 + 1 ACUGCGGAGUGCCAGGACCACAGUGAAGACCAACCUCAAACUGGUCCUGUCC------UCCGUCCUCCGUCGAUUUCCUCACUCA------------C ...((((((.(.((((((((...(((.(......))))...)))))))).))------)))))......................------------. ( -25.70) >consensus ACUGCGGAGUGCCAGGACCACAGUGAAGACCGACCUCAAACUGGUCCUGUCCCCCA______UCCUCCGUCGAUUUCCUCACUCCGGACCAUCAAUCC ....(((((((.((((((((...(((.(.....).)))...))))))))...............(......).......)))))))............ (-20.09 = -20.90 + 0.81)

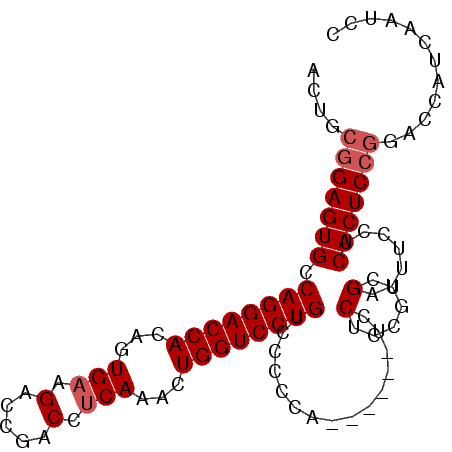
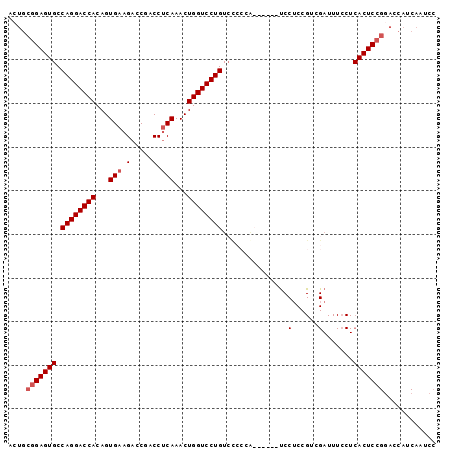
| Location | 16,877,869 – 16,877,961 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.85 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16877869 92 - 20766785 GGAUUGAUGCUACGGAGUGAGGAAAUCGACGGCGGA------CGGGGUACAGGACCAGUUUGAGGUCGGUCUUCACUGUGGUCCUGGCACUCCGCAGU ........(((.(((((((.....(((..((.....------)).))).((((((((...(((((.....)))))...)))))))).))))))).))) ( -35.20) >DroSec_CAF1 28223 77 - 1 GGAUUGAUGGUCCGGAGUG---------------------AGUGGGGGACAGGACCAGUUUGUGGUCGGUCUUCACUGUGGUCCUGGCACUCCGCAGU ((((.....))))......---------------------.((((((..((((((((....((((.......))))..))))))))...))))))... ( -31.90) >DroSim_CAF1 28531 94 - 1 GGAUUGAUGGUCCGGAGUGAGCAAAUCGGCGGAGGG----AGUGGGAGACAGGACCAGUUUGAGGUCGGUCUUCACUGUGGUCCUGGCACUCCGCAGU ((((.....))))((((((......((.((......----.)).))...((((((((...(((((.....)))))...)))))))).))))))..... ( -35.50) >DroEre_CAF1 28591 80 - 1 G------------UGAGUGAGGAAAUCGACGGAGGACGGA------GGACAGGACCAGUUUGAGGUUGGUCUUCACUGUGGUCCUGGCACUCCGCAGU (------------((((((......((..((.....))..------)).((((((((...(((((.....)))))...)))))))).)))).)))... ( -29.30) >consensus GGAUUGAUGGUCCGGAGUGAGGAAAUCGACGGAGGA______UGGGGGACAGGACCAGUUUGAGGUCGGUCUUCACUGUGGUCCUGGCACUCCGCAGU ............(((((((......((......))..............((((((((...(((((.....)))))...)))))))).))))))).... (-24.98 = -25.85 + 0.87)

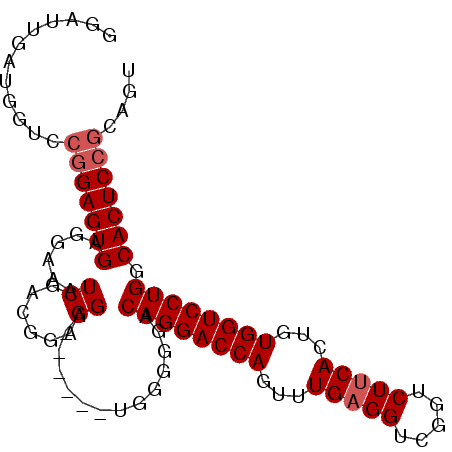
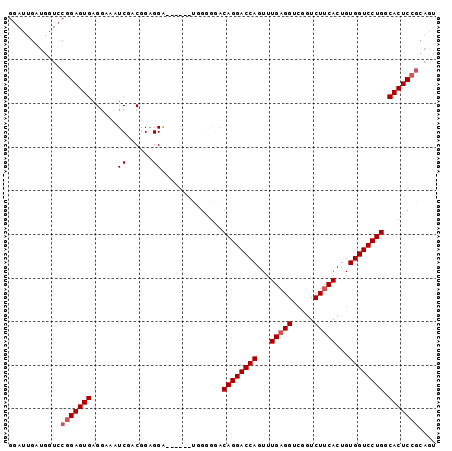
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:53 2006