
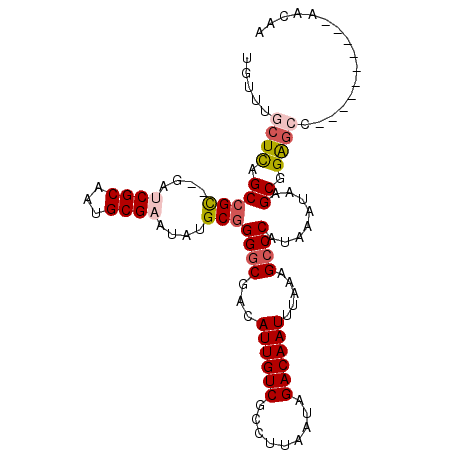
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,865,900 – 16,866,003 |
| Length | 103 |
| Max. P | 0.525211 |

| Location | 16,865,900 – 16,866,003 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16865900 103 + 20766785 UGUUUGCUCAGCCGC--GAUCGCAAUGCGAAUAUGCGGGGCGACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCGGAACAAUAAACAAUAAACAA ((((((.....((((--...((((((....)).))))((((.(.((((((.........)))))).)...))))........)))).....))))))........ ( -28.30) >DroSec_CAF1 16302 93 + 1 UGUUUGCUCAGCCGC--GAACGCAAUGCGAAUAUGCGGGGCGACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCGGAGCC----------AACAA ((((.((((.(((((--(..(((...)))....))))((((.(.((((((.........)))))).)...))))........)).)))).----------)))). ( -30.20) >DroSim_CAF1 16417 93 + 1 UGUUUGCUCAGCCGC--GAACGCAAUGCGAAUAUGCGGGGCGACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCGGAGCC----------AACAA ((((.((((.(((((--(..(((...)))....))))((((.(.((((((.........)))))).)...))))........)).)))).----------)))). ( -30.20) >DroEre_CAF1 16569 93 + 1 UGUUUGCUCAGCCGC--GAUCGCAAUGCGAAUAUGCGGGGCGACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCGGAGCC----------AACAA ((((.((((.(((((--(.((((...))))...))))((((.(.((((((.........)))))).)...))))........)).)))).----------)))). ( -32.20) >DroYak_CAF1 16576 93 + 1 UGUUUUCUCAGCCGC--GAUCGCAAUGCGAAUAUGCGGGGCGACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCGGAGCC----------AACAA ((((..(((.(((((--(.((((...))))...))))((((.(.((((((.........)))))).)...))))........)).)))..----------)))). ( -28.00) >DroAna_CAF1 19844 93 + 1 UGUUUUCUUAGCGGUGAGAUCGCAAUGCGAA--UGCGGGACAACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCAGGGCC----------AACAU (((((..((((.(((....(((((.......--)))))((((....))))))))))).))))).......((((...........)))).----------..... ( -19.50) >consensus UGUUUGCUCAGCCGC__GAUCGCAAUGCGAAUAUGCGGGGCGACAUUGUCGCCUUAAUAGACAAUUUAAAGCCCAUAAAUAAGCGGAGCC__________AACAA .....((((.(((((....((((...))))....)))((((...((((((.........)))))).....))))........)).))))................ (-21.75 = -22.50 + 0.75)


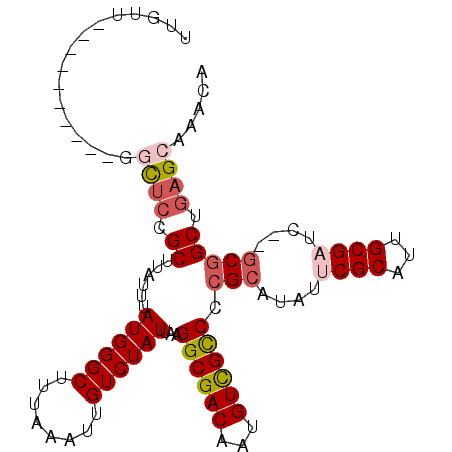
| Location | 16,865,900 – 16,866,003 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -22.67 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16865900 103 - 20766785 UUGUUUAUUGUUUAUUGUUCCGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUCGCCCCGCAUAUUCGCAUUGCGAUC--GCGGCUGAGCAAACA ..............((((((.((......((((((........))))))...((((((...)))))).(((....((((...))))..--))))).))))))... ( -28.60) >DroSec_CAF1 16302 93 - 1 UUGUU----------GGCUCCGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUCGCCCCGCAUAUUCGCAUUGCGUUC--GCGGCUGAGCAAACA .((((----------.(((((((((....((((((........))))))..))))).....(((((..((((.........))))...--))))).)))).)))) ( -30.00) >DroSim_CAF1 16417 93 - 1 UUGUU----------GGCUCCGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUCGCCCCGCAUAUUCGCAUUGCGUUC--GCGGCUGAGCAAACA .((((----------.(((((((((....((((((........))))))..))))).....(((((..((((.........))))...--))))).)))).)))) ( -30.00) >DroEre_CAF1 16569 93 - 1 UUGUU----------GGCUCCGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUCGCCCCGCAUAUUCGCAUUGCGAUC--GCGGCUGAGCAAACA .((((----------.((((.((......((((((........))))))...((((((...)))))).(((....((((...))))..--))))).)))).)))) ( -31.20) >DroYak_CAF1 16576 93 - 1 UUGUU----------GGCUCCGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUCGCCCCGCAUAUUCGCAUUGCGAUC--GCGGCUGAGAAAACA .((((----------..(((.((......((((((........))))))...((((((...)))))).(((....((((...))))..--))))).)))..)))) ( -29.20) >DroAna_CAF1 19844 93 - 1 AUGUU----------GGCCCUGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUUGUCCCGCA--UUCGCAUUGCGAUCUCACCGCUAAGAAAACA .(.((----------(((..(((......((((((........))))))...((.((((....)))))))))--.((((...)))).......))))).)..... ( -20.80) >consensus UUGUU__________GGCUCCGCUUAUUUAUGGGCUUUAAAUUGUCUAUUAAGGCGACAAUGUCGCCCCGCAUAUUCGCAUUGCGAUC__GCGGCUGAGCAAACA ................((((.((......((((((........))))))...((((((...)))))).(((....((((...))))....))))).))))..... (-22.67 = -23.58 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:50 2006