
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,864,983 – 16,865,091 |
| Length | 108 |
| Max. P | 0.780974 |

| Location | 16,864,983 – 16,865,091 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
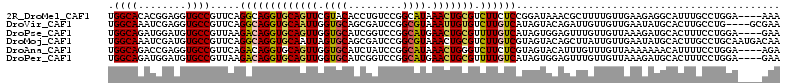
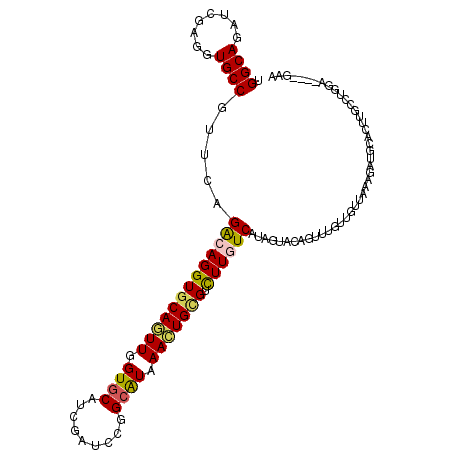
>2R_DroMel_CAF1 16864983 108 + 20766785 UGGCACACGGAGGUGCCGUUCAGGCAGGUGCAGUUCGUACACCUGUCCGGCAUAAACUGCGUCUUCUCCGGAUAAACGCUUUUGUUGAAGAGGCAUUUGCCUGGA----AAA .((((...(...((((((..((((...((((.....)))).))))..))))))...)(((.(((((..((((........))))..))))).)))..))))....----... ( -40.50) >DroVir_CAF1 26931 108 + 1 UGGCAAAUCGAGGUGCCGUUCAGGCAGGUGCAAUUGGUGCAGCGAUCCGGCGUAAAUUGUGUCUUGUCAUAGUACAGAUUGUUGUUGAAUAUGCACUUGCCUG----GCGAA .((((........))))((.((((((((((((......(((((((((.(......((((((......)))))).).)))))))))......))))))))))))----))... ( -41.30) >DroPse_CAF1 15092 108 + 1 UGGCAGAUGGAUGUGCCGUUAAGACAGGUGCAGUUGGUGCAUCGGUCCGGCAUGAACUGCGUUUUGUCAUAGUGGAGUUUGUUGUUAAAGAUGCACUUUCCUGGA----GAA .((((.(....).)))).....((((..(((((((.((((.........)))).)))))))..).))).(((..((((.((((......)))).))))..)))..----... ( -32.20) >DroMoj_CAF1 22289 112 + 1 UGGCAAAUCGAUGUGCCGUUCAGGCAGGUGCAAUUAGUGCAGCGAUCCGGCGUAAACUGCGUCUUGUCGUAGUACAGCUUAUUGUUGAAUAUGCACUUGCCUGCAAUGACAA .((((........))))(((((((((((((((....((((.(((((..(((((.....)))))..))))).))))(((.....))).....)))))))))))).)))..... ( -45.30) >DroAna_CAF1 19131 108 + 1 UGGCAGACCGAGGUGCCGUUCAGACAGGUGCAGUUGGUGCAUCUAUCCGGCAUAAACUGGGUCUUCUCGUAGUACAUUUGUUUGUUAAAAAAACAUUUUCCUGGA----AGA ....(((((.((((((((....((.(((((((.....))))))).))))))))...)).)))))(((..(((......(((((.......))))).....)))..----))) ( -30.40) >DroPer_CAF1 16618 108 + 1 UGGCAGAUGGAUGUGCCGUUAAGACAGGUGCAGUUGGUGCAUCGGUCCGGCAUGAACUGCGUUUUGUCAUAGUGGAGUUUGUUGUUAAAGAUGCACUUUCCUGGA----GAA .((((.(....).)))).....((((..(((((((.((((.........)))).)))))))..).))).(((..((((.((((......)))).))))..)))..----... ( -32.20) >consensus UGGCAGAUCGAGGUGCCGUUCAGACAGGUGCAGUUGGUGCAUCGAUCCGGCAUAAACUGCGUCUUGUCAUAGUACAGUUUGUUGUUAAAGAUGCACUUGCCUGGA____GAA .((((........)))).....(((((((((((((.((((.........)))).))))))).))))))............................................ (-20.42 = -20.23 + -0.19)
| Location | 16,864,983 – 16,865,091 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.39 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
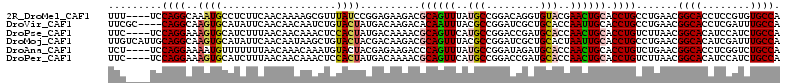
>2R_DroMel_CAF1 16864983 108 - 20766785 UUU----UCCAGGCAAAUGCCUCUUCAACAAAAGCGUUUAUCCGGAGAAGACGCAGUUUAUGCCGGACAGGUGUACGAACUGCACCUGCCUGAACGGCACCUCCGUGUGCCA ...----..(((((((((((.............)))))).(((((...((((...))))...))))).(((((((.....))))))))))))...(((((......))))). ( -36.92) >DroVir_CAF1 26931 108 - 1 UUCGC----CAGGCAAGUGCAUAUUCAACAACAAUCUGUACUAUGACAAGACACAAUUUACGCCGGAUCGCUGCACCAAUUGCACCUGCCUGAACGGCACCUCGAUUUGCCA .....----((((((.(((((.......((.(.(((((.........................))))).).)).......))))).))))))...((((........)))). ( -26.05) >DroPse_CAF1 15092 108 - 1 UUC----UCCAGGAAAGUGCAUCUUUAACAACAAACUCCACUAUGACAAAACGCAGUUCAUGCCGGACCGAUGCACCAACUGCACCUGUCUUAACGGCACAUCCAUCUGCCA ...----..((((..((((...................))))..........((((((..(((.........)))..)))))).)))).......((((........)))). ( -21.31) >DroMoj_CAF1 22289 112 - 1 UUGUCAUUGCAGGCAAGUGCAUAUUCAACAAUAAGCUGUACUACGACAAGACGCAGUUUACGCCGGAUCGCUGCACUAAUUGCACCUGCCUGAACGGCACAUCGAUUUGCCA .........((((((.(((((..........((((((((.((......))..)))))))).((.((....))))......))))).))))))...((((........)))). ( -31.40) >DroAna_CAF1 19131 108 - 1 UCU----UCCAGGAAAAUGUUUUUUUAACAAACAAAUGUACUACGAGAAGACCCAGUUUAUGCCGGAUAGAUGCACCAACUGCACCUGUCUGAACGGCACCUCGGUCUGCCA (((----(..((.....(((((.......)))))......))..))))(((((.((....((((((((((.((((.....)))).)))))....))))).)).))))).... ( -26.10) >DroPer_CAF1 16618 108 - 1 UUC----UCCAGGAAAGUGCAUCUUUAACAACAAACUCCACUAUGACAAAACGCAGUUCAUGCCGGACCGAUGCACCAACUGCACCUGUCUUAACGGCACAUCCAUCUGCCA ...----..((((..((((...................))))..........((((((..(((.........)))..)))))).)))).......((((........)))). ( -21.31) >consensus UUC____UCCAGGAAAGUGCAUCUUCAACAACAAACUGUACUAUGACAAGACGCAGUUUAUGCCGGACCGAUGCACCAACUGCACCUGCCUGAACGGCACAUCCAUCUGCCA .........((((..((((...................))))..........((((((..(((.........)))..)))))).)))).......((((........)))). (-15.34 = -15.39 + 0.06)
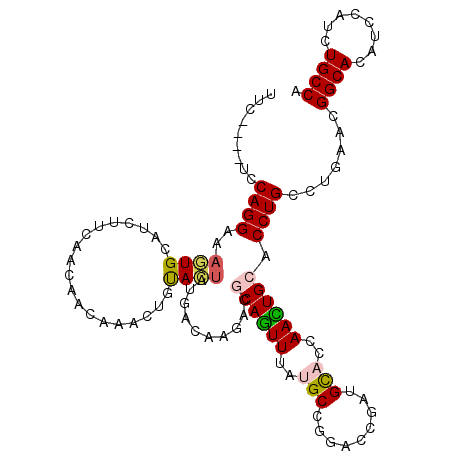
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:47 2006