
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,857,879 – 16,858,011 |
| Length | 132 |
| Max. P | 0.988012 |

| Location | 16,857,879 – 16,857,990 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -25.61 |
| Energy contribution | -26.42 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16857879 111 - 20766785 AUAUUCCAGCUGAUUUUCCAGUUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAAUGGCGUCAGACAUAUCGCGUGCCGAACAACUGGAGUAAGU .((((((((((((((..((((.(((((((.....)))))))...))))...))).))).......(((((...(((..........))).))))).....))))))))... ( -37.80) >DroSec_CAF1 8306 111 - 1 GUAUUCCAGCUGACUUUCCAGCUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAAUGGCGUCAGACAUAUCGCGUGCCGAACAACUGGAGUAAGU .(((((((((((..((.((((.(((((((.....)))))))...))))...))..))).......(((((...(((..........))).))))).....))))))))... ( -35.40) >DroSim_CAF1 8349 111 - 1 GUACUCCAGCUGAUUUUCCAGCUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAGUGGAGUCAGACAUAUCGCGUGCCGAACAACUGGAGUAAGU .((((((((((((((..((((.(((((((.....)))))))...))))...))).))).......(((((((((.((.....)).)))).))))).....))))))))... ( -38.70) >DroEre_CAF1 8482 111 - 1 GUAUUCCAGCUGAUUUUCCAGUUGGUCCUGGCAAACGAUUAUUCCUGGUGUAAUCCACAAUUAUGCGGCAACGGAGUGAAACAUAUCGCGUGCCGAAAUACUGGAGUAAGU ((((((((((((......)))))))...(((((..((((((((((..(((((((.....)))))))(....))))))))......)))..))))).))))).......... ( -32.30) >DroYak_CAF1 8395 111 - 1 GUAUUCCAGCUGAUUUUCCAGUUGGUCCUGGCUACGGAUUAUUCCUGGUGUAAUCCAGAGUUAUGCGGCAAUGGUGUGAAACAUAUCGCGUGCCGAAAUACUGGAGUAAGU .((((((((((((((..((((.((((((.......))))))...))))...))).))).......(((((.(((((((...)))))))..))))).....))))))))... ( -35.90) >DroAna_CAF1 11762 111 - 1 AUUUUCCAGCUGAUGGUGCAGUUGGUCCUGGCCAAGGACUUCAGCUGGUGUGAUCCCAGUCUGUGUGAGCCCACCCAGAGGCACAUUGCCUGCCGGAAUACUGGUGUAAGU .....((((((((.(((.(..(((((....)))))).)))))))))))....(..((((((((.(((....))).))))((((.......))))......))))..).... ( -38.80) >consensus GUAUUCCAGCUGAUUUUCCAGUUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAAUGGCGUCAGACAUAUCGCGUGCCGAAAAACUGGAGUAAGU .((((((((..((((..((((.(((((((.....)))))))...))))...))))......(((((((..(((........))).)))))))........))))))))... (-25.61 = -26.42 + 0.81)

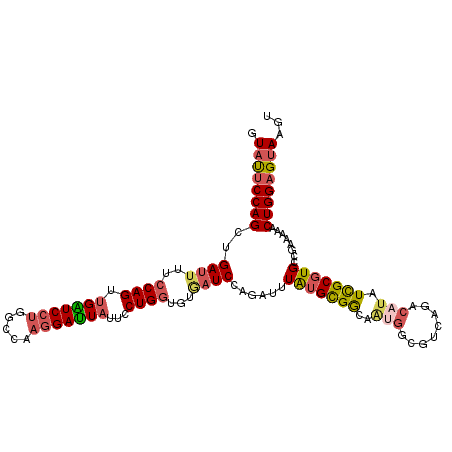
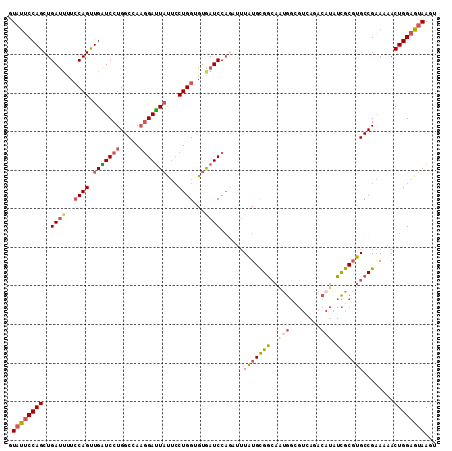
| Location | 16,857,919 – 16,858,011 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16857919 92 + 20766785 UUGCCGCAUAAAUCUGGAUCACACCAGGAAUAAUCCUUGGCCAGGAUCAACUGGAAAAUCAGCUGGAAUAUGGCCAAGCAGA-------------------UCAAAGGCGA (((((.......(((((......))))).......((((((((...(((.(((......))).)))....))))))))....-------------------.....))))) ( -28.90) >DroSec_CAF1 8346 91 + 1 UUGCCGCAUAAAUCUGGAUCACACCAGGAAUAAUCCUUGGCCAGGAUCAGCUGGAAAGUCAGCUGGAAUACGGCCAGGCAGA-------------------UCAAAGGCG- ..(((.......(((((......))))).......(((((((....((((((((....)))))))).....)))))))....-------------------.....))).- ( -32.40) >DroSim_CAF1 8389 91 + 1 CUGCCGCAUAAAUCUGGAUCACACCAGGAAUAAUCCUUGGCCAGGAUCAGCUGGAAAAUCAGCUGGAGUACGGCCAGGCAGA-------------------UCAAAGGCG- (((((.......(((((......))))).........(((((....(((((((......))))))).....)))))))))).-------------------.........- ( -32.70) >DroEre_CAF1 8522 103 + 1 UUGCCGCAUAAUUGUGGAUUACACCAGGAAUAAUCGUUUGCCAGGACCAACUGGAAAAUCAGCUGGAAUACGGCCAGGCAGACACCUCCACGG-------AUCAAAGGCA- .((((......(((((((.........((....))(((((((....((....)).......((((.....))))..)))))))...)))))))-------......))))- ( -30.50) >DroYak_CAF1 8435 103 + 1 UUGCCGCAUAACUCUGGAUUACACCAGGAAUAAUCCGUAGCCAGGACCAACUGGAAAAUCAGCUGGAAUACAGCCAGGAAGACACUUCCACGA-------CUCAAAGGCG- ..(((.......(((((......))))).......(((..((((......)))).......((((.....))))..(((((...)))))))).-------......))).- ( -28.40) >DroAna_CAF1 11802 109 + 1 GGCUCACACAGACUGGGAUCACACCAGCUGAAGUCCUUGGCCAGGACCAACUGCACCAUCAGCUGGAAAAUGGCCACGCAGGCAG-UCCAGGAAAUGACCAUCGAAGACA- ((.(((..(((.((((.......)))))))...((((((((((...(((.(((......))).)))....))))))....((...-.))))))..)))))..........- ( -32.40) >consensus UUGCCGCAUAAAUCUGGAUCACACCAGGAAUAAUCCUUGGCCAGGACCAACUGGAAAAUCAGCUGGAAUACGGCCAGGCAGA___________________UCAAAGGCG_ ..(((.......(((((......))))).......(((((((....(((.(((......))).))).....)))))))............................))).. (-18.94 = -19.88 + 0.95)

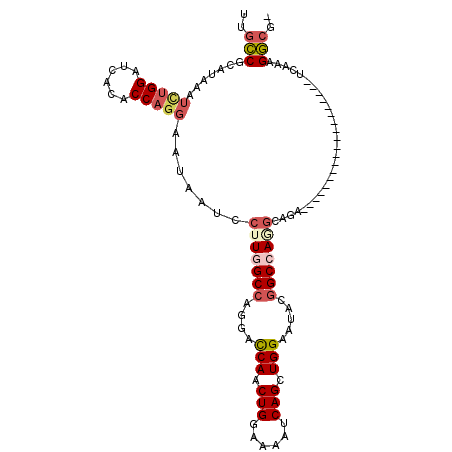
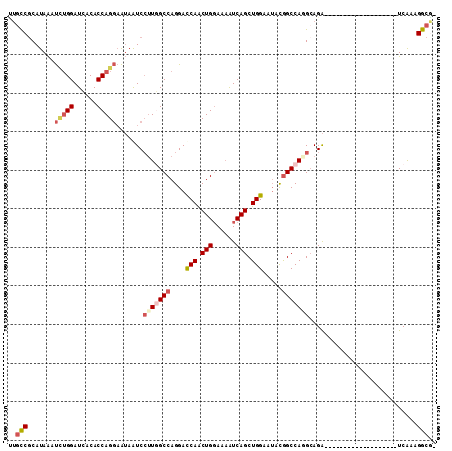
| Location | 16,857,919 – 16,858,011 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -25.43 |
| Energy contribution | -24.93 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16857919 92 - 20766785 UCGCCUUUGA-------------------UCUGCUUGGCCAUAUUCCAGCUGAUUUUCCAGUUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAA ..(((...((-------------------((((((((((((.....((((((......))))))....))))))))((((((.......)))))))))))).....))).. ( -34.00) >DroSec_CAF1 8346 91 - 1 -CGCCUUUGA-------------------UCUGCCUGGCCGUAUUCCAGCUGACUUUCCAGCUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAA -.(((...((-------------------(((...((((((.....((((((......))))))....)))))).(((((((.......)))))))))))).....))).. ( -33.70) >DroSim_CAF1 8389 91 - 1 -CGCCUUUGA-------------------UCUGCCUGGCCGUACUCCAGCUGAUUUUCCAGCUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAG -.(((...((-------------------(((...((((((.....((((((......))))))....)))))).(((((((.......)))))))))))).....))).. ( -33.70) >DroEre_CAF1 8522 103 - 1 -UGCCUUUGAU-------CCGUGGAGGUGUCUGCCUGGCCGUAUUCCAGCUGAUUUUCCAGUUGGUCCUGGCAAACGAUUAUUCCUGGUGUAAUCCACAAUUAUGCGGCAA -((((......-------.((.((((.(((.((((.((.......(((((((......))))))).)).)))).)))....))))))(((((((.....))))))))))). ( -32.10) >DroYak_CAF1 8435 103 - 1 -CGCCUUUGAG-------UCGUGGAAGUGUCUUCCUGGCUGUAUUCCAGCUGAUUUUCCAGUUGGUCCUGGCUACGGAUUAUUCCUGGUGUAAUCCAGAGUUAUGCGGCAA -.(((....((-------(((.(((((...))))))))))(((..(((((((......)))))))...(((((..(((((((.......)))))))..))))))))))).. ( -35.80) >DroAna_CAF1 11802 109 - 1 -UGUCUUCGAUGGUCAUUUCCUGGA-CUGCCUGCGUGGCCAUUUUCCAGCUGAUGGUGCAGUUGGUCCUGGCCAAGGACUUCAGCUGGUGUGAUCCCAGUCUGUGUGAGCC -.(((((...((((((......(((-((..((((...((((((........))))))))))..))))))))))))))))..(((((((.(....))))).)))........ ( -37.30) >consensus _CGCCUUUGA___________________UCUGCCUGGCCGUAUUCCAGCUGAUUUUCCAGUUGAUCCUGGCCAAGGAUUAUUCCUGGUGUGAUCCAGAUUUAUGCGGCAA ..(((..............................((((((.....((((((......))))))....)))))).(((((((.......)))))))..........))).. (-25.43 = -24.93 + -0.49)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:43 2006