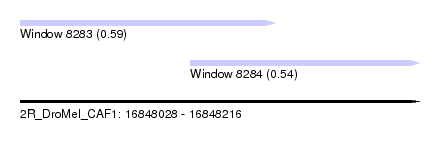
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,848,028 – 16,848,216 |
| Length | 188 |
| Max. P | 0.593999 |

| Location | 16,848,028 – 16,848,148 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -27.28 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16848028 120 + 20766785 CAAAACACAAAGGGACUAUCGAAUUCGGUGACCAUGCAGCAGAUUCAACAAGUUAUGCGAAGUGGCCAGCAGGGCACGUUAGCCACCACCAAUCUGGUGUUGGGUAAAACAAGCGUGGGA ................(((((....))))).((((((.(((((((.....)))).)))...(((.((....)).))).(((.(((.((((.....)))).))).))).....)))))).. ( -34.90) >DroPse_CAF1 17647 120 + 1 CAAAACACCAAGACGCUGCCCAGCUCGGUGACGGUGCAGCAGUUCCAACAGGUCAUGCGCAGUGGGCAGCAGGGGACGUUGGCCACCACGAAUCUGGUGCUGGGCAAGACUAGUGUCGGA .......((..(((((((((((((((((...((.(((.(((...((....))...))))))((((.((((.(....))))).))))..))...)))).)))))))......)))))))). ( -49.70) >DroWil_CAF1 18172 117 + 1 CAAAACACGAAAGCUUUGCCCAGUUCGGUGACGGUUCAGCAAAUCCAACAGGUAAUGCGUAGUGGUCAACAGGGAACAUUGGCCAC---CAAUCUGGUGCUGGGCAAAACUAGUGUCGGA ....((((...((.(((((((((((((....))).((((((...((....))...)))...((((((((..(....).))))))))---.....))).)))))))))).)).)))).... ( -40.20) >DroYak_CAF1 16276 120 + 1 CAAAACACAAAAGCACUGCCGAAUUCGGUCACCGUGCAGCAGAUUCAACAAGUGAUGCGUAGUGGCCAGCAGGGUACGUUAGCCACCACCAAUCUGGUGUUGGGUAAGACGAGCGUGGGA .....(((.......((((.(....)((((((..((((.((...........)).))))..)))))).)))).((.(((((.(((.((((.....)))).))).)..)))).)))))... ( -37.70) >DroAna_CAF1 16782 120 + 1 CAGAACACGAAGGCCUUGCCCAAUUCGGUGACUGUUCAGCAAAUCCAGCAGGUGAUGCGCAGUGGCCAACAGGGCACUCUGGCCACCACGAACCUGGUGCUGGGCAAGACCAGCGUGGGU .....((((..((.((((((((....(.(((....))).).....((.(((((....((..(((((((..((....)).)))))))..)).))))).)).)))))))).))..))))... ( -50.80) >DroPer_CAF1 17562 120 + 1 CAAAACACCAAGACGCUGCCCAGCUCGGUGACGGUGCAGCAGUUCCAACAGGUUAUGCGCAGUGGGCAGCAGGGGACGUUGGCCACCACGAAUCUGGUGCUGGGCAAGACUAGUGUCGGA .......((..(((((((((((((((((...((.(((.(((...((....))...))))))((((.((((.(....))))).))))..))...)))).)))))))......)))))))). ( -49.70) >consensus CAAAACACAAAGGCACUGCCCAAUUCGGUGACGGUGCAGCAGAUCCAACAGGUGAUGCGCAGUGGCCAGCAGGGCACGUUGGCCACCACCAAUCUGGUGCUGGGCAAGACUAGCGUCGGA ................((((((((((((..........(((...((....))...)))...(((((((((.(....)))))))))).......)))).)))))))).............. (-27.28 = -27.82 + 0.53)

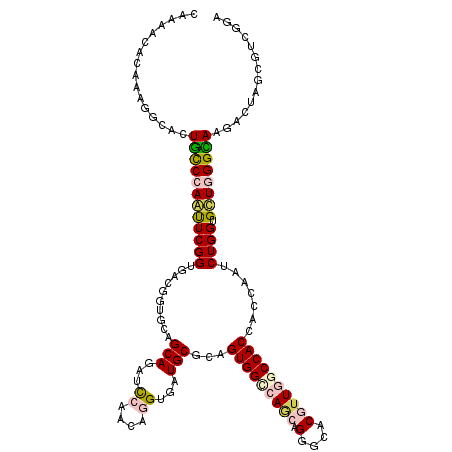
| Location | 16,848,108 – 16,848,216 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16848108 108 + 20766785 AGCCACCACCAAUCUGGUGUUGGGUAAAACAAGCGUGGGACGGGUAAUCCCCGUGUCGGUGGCUUCACAGGCCAACCAGCGUCAAACCAUUCAGGUAGGUUU-CUUGUU ..(((.((((.....)))).)))....(((((((((((.(((((.....))))).))(((((((.....)))).))).)))..(((((.........)))))-)))))) ( -37.20) >DroPse_CAF1 17727 108 + 1 GGCCACCACGAAUCUGGUGCUGGGCAAGACUAGUGUCGGACGGGUCAUACCCGUGUCGGUGGCCUCGCAGGCCAAUCAGCGGCAGACGAUCCAGGUAGGUUA-CCUAUC ((((((((((..(((((..((((......))))..)))))((((.....))))))).)))))))(((...(((.......)))...)))....((((((...-)))))) ( -46.90) >DroWil_CAF1 18252 105 + 1 GGCCAC---CAAUCUGGUGCUGGGCAAAACUAGUGUCGGACGUGUAAUACCCGUUUCGGUGGCCUCGCAGGCCAAUCAACGUCAGACCAUACAGGUAAGAUC-CCCACA ((((((---(..(((((..((((......))))..)))))((.(.....).))....)))))))................(((..(((.....)))..))).-...... ( -33.00) >DroYak_CAF1 16356 108 + 1 AGCCACCACCAAUCUGGUGUUGGGUAAGACGAGCGUGGGACGGGUGAUUCCCGUGUCGGUGGCUUCACAGGCCAACCAGCGUCAAACCAUUCAGGUGGGUUA-UUUGUU ..((((((((.....)))....(((..((((.....((.(((((.....))))).))(((((((.....)))).)))..))))..))).....)))))....-...... ( -41.10) >DroAna_CAF1 16862 109 + 1 GGCCACCACGAACCUGGUGCUGGGCAAGACCAGCGUGGGUCGGGUGAUUCCCGUCUCCGUGGCAUCACAGGCCAACCAACGACAGACUAUUCAGGUGGGUUCUAUAGCU ..(((((.......(((((((((......)))))...((((..(((((.((((....)).)).))))).)))).))))..((........)).)))))........... ( -38.10) >DroPer_CAF1 17642 108 + 1 GGCCACCACGAAUCUGGUGCUGGGCAAGACUAGUGUCGGACGGGUCAUACCCGUGUCGGUGGCCUCGCAGGCCAAUCAGCGGCAGACGAUCCAGGUAGGUUA-CCUAUC ((((((((((..(((((..((((......))))..)))))((((.....))))))).)))))))(((...(((.......)))...)))....((((((...-)))))) ( -46.90) >consensus GGCCACCACCAAUCUGGUGCUGGGCAAGACUAGCGUCGGACGGGUAAUACCCGUGUCGGUGGCCUCACAGGCCAACCAGCGUCAGACCAUUCAGGUAGGUUA_CCUAUU ....(((....((((((.(((((......)))))((((.(((((.....)))))...(((((((.....)))).))).........)))))))))).)))......... (-27.40 = -27.40 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:34 2006