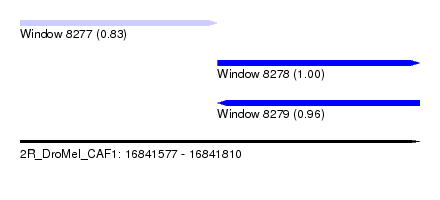
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,841,577 – 16,841,810 |
| Length | 233 |
| Max. P | 0.999485 |

| Location | 16,841,577 – 16,841,692 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -19.98 |
| Energy contribution | -19.46 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16841577 115 + 20766785 UGGCACAGAGAAA-AUCUUCCUUUGGAAAGUAGACUUUCAGACAUAGUUGAGGCGACA-UAACCCACGCCACGAUUGUAUCACUCAAAGUCUAAUAUCUGAAAAAUCUUCGAUCAUA .....((((....-...((((...))))..((((((((..((.(((((((.((((...-.......)))).))))))).))....))))))))...))))................. ( -25.30) >DroSec_CAF1 10498 115 + 1 UGGCACAGAGAAA-AUCUUCCUUUGGAAAAUAGACCUUCAGACAUAGUUGAGGCGACA-UAACCUACGCCACGAUUGUAUCACUCAAAGUCUAAUAUCUGAAAAAUCUUCGAUCAUA .....((((....-...((((...))))..(((((.....((.(((((((.((((...-.......)))).))))))).)).......)))))...))))................. ( -21.50) >DroSim_CAF1 9700 115 + 1 UGGCACAGAAAAA-AUCUUCCUUUGGAAAAUAGACCUUCAGACAUAGUUGAGGCGAAA-UAACCCACGCCACGAUUGUAUCACUCAAAGUCUAAUAUUUGAAAAAUCUUCGAUCAUA .....((((....-...((((...))))..(((((.....((.(((((((.((((...-.......)))).))))))).)).......)))))...))))................. ( -18.90) >DroEre_CAF1 11148 110 + 1 CGGCACAGAAAAAUAUCUUCCUUUGGAAAGUAGACCUUCAGACAUAGUCGAGGUGACC-UAAUCCACGCCACGAUUGUAUCACUCAAAGUCUAAUAUCUGAAUAU------AUUAUA .....((((........((((...))))..(((((.....((.(((((((.((((...-.......)))).))))))).)).......)))))...)))).....------...... ( -21.60) >DroYak_CAF1 9646 116 + 1 CGGCACAGAAAAA-AUCUUCCUUUGGAAAGUAAACUUUCAGACAUAGUCGAGGCGACCUUAAUCAACGCCACGAUUGUAUCACUCAAAGUCUAAUAUCUAAAAAAUCACCAUUUAUA .((....(((...-...)))((((((((((....))))).((.(((((((.((((...........)))).))))))).))...)))))...................))....... ( -24.00) >consensus UGGCACAGAAAAA_AUCUUCCUUUGGAAAGUAGACCUUCAGACAUAGUUGAGGCGACA_UAACCCACGCCACGAUUGUAUCACUCAAAGUCUAAUAUCUGAAAAAUCUUCGAUCAUA .((...(((......))).))((((((...(((((.....((.(((((((.((((...........)))).))))))).)).......)))))...))))))............... (-19.98 = -19.46 + -0.52)

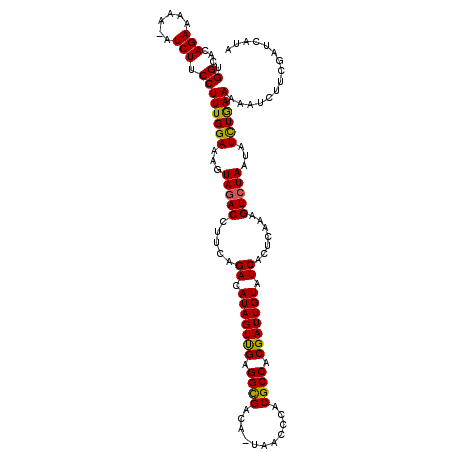
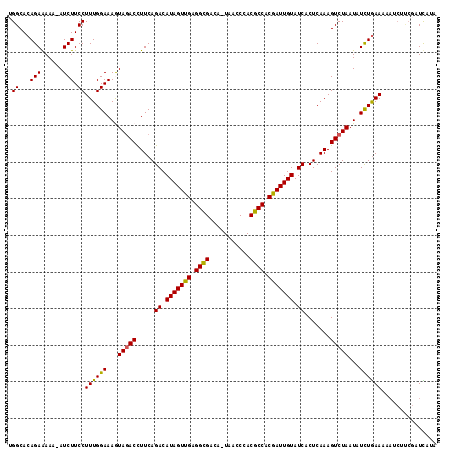
| Location | 16,841,692 – 16,841,810 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -26.68 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16841692 118 + 20766785 CUAUUUU-UUU-UACAACGCCAAACUAGUUUUUAUUGCGGCAUCGCUUGCAACUCAUUGAAUUCGUUUUUCUAAUCGAAUAAAAUGAAGUGAGUGUUGCCAAGAGCAAUACACCUUUUCG .......-...-....................((((((((((.((((..(...(((((..(((((.((....)).)))))..))))).)..)))).))))....)))))).......... ( -26.40) >DroSec_CAF1 10613 118 + 1 CUGUUCU-UUU-UACAAUGCCAAAUUAGUUUUUAUUGCGGCAUCGCUUGCAACUCAUUGAAUUCGUUUUUCUAAUCGAAUCCAAUGAAGUGAGUGAUGCCAAGAGCAAUACAUCUUUUCG .((((((-(..-..(((((..(((.....)))))))).(((((((((..(...((((((.(((((.((....)).))))).)))))).)..))))))))))))))))............. ( -36.00) >DroSim_CAF1 9815 118 + 1 CUGUUCU-UUU-UACAAUGCCAAAUUAGUUUUUAUUGCGGCAUCGCUUGCAACUCAUUGAAUUCGUUUUUCUAAUCGAAUCCAAUGAAGUGAGUGAUGCCAAGAGCAAUACAUCUUUUCG .((((((-(..-..(((((..(((.....)))))))).(((((((((..(...((((((.(((((.((....)).))))).)))))).)..))))))))))))))))............. ( -36.00) >DroEre_CAF1 11258 119 + 1 CUAUUUU-UUUUGACAACGUAGAAAUAGUUUUAAUUGCGGCAUCACUUGCAACUUAAUGAAUUCGUAUUUCUAAUCGAAUACAACAAAGUAAGUGUUGCCAAGAGUAAUACACCUCUACG .......-.........((((((....((.(((.((..((((.(((((((........(....)((((((......))))))......))))))).))))..)).))).))...)))))) ( -26.30) >DroYak_CAF1 9762 114 + 1 CUAUUUUUUUUUGACAACG-AAAACUAGUUUUUACUGCGGCAACACUUGCAACA--UUGAAUUCGUAAUUGUAAUCGAAUCCAAUGAAGUGAGUGUUGCCAAGAGUAAU---CCUUUUCG .................((-((((.......(((((..(((((((((..(..((--(((.(((((..........))))).)))))..)..)))))))))...))))).---..)))))) ( -34.90) >consensus CUAUUUU_UUU_UACAACGCCAAACUAGUUUUUAUUGCGGCAUCGCUUGCAACUCAUUGAAUUCGUUUUUCUAAUCGAAUCCAAUGAAGUGAGUGUUGCCAAGAGCAAUACACCUUUUCG .((((((....................((.......))((((((((((((...((((((.(((((..........))))).)))))).)))))))))))).))))))............. (-26.68 = -27.12 + 0.44)

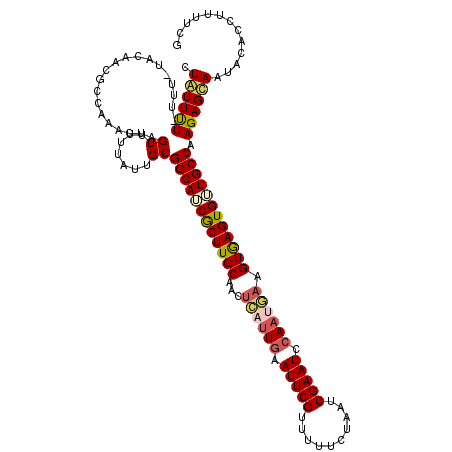
| Location | 16,841,692 – 16,841,810 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
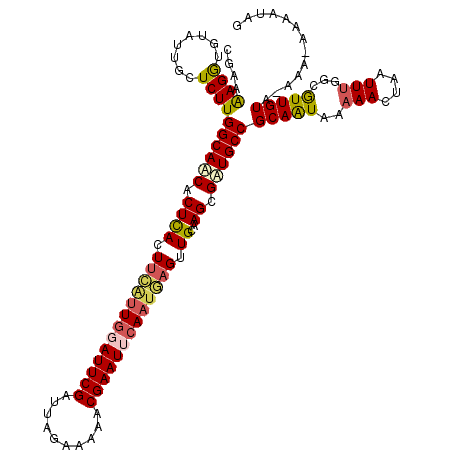
>2R_DroMel_CAF1 16841692 118 - 20766785 CGAAAAGGUGUAUUGCUCUUGGCAACACUCACUUCAUUUUAUUCGAUUAGAAAAACGAAUUCAAUGAGUUGCAAGCGAUGCCGCAAUAAAAACUAGUUUGGCGUUGUA-AAA-AAAAUAG ......(((((..(((.....))))))))((.((((((..(((((.((....)).)))))..)))))).))...(((((((((.(((........)))))))))))).-...-....... ( -25.30) >DroSec_CAF1 10613 118 - 1 CGAAAAGAUGUAUUGCUCUUGGCAUCACUCACUUCAUUGGAUUCGAUUAGAAAAACGAAUUCAAUGAGUUGCAAGCGAUGCCGCAAUAAAAACUAAUUUGGCAUUGUA-AAA-AGAACAG ......((((((((((....((((((.((((.(((((((((((((.((....)).))))))))))))).))..)).))))))))))).....(......))))))...-...-....... ( -31.60) >DroSim_CAF1 9815 118 - 1 CGAAAAGAUGUAUUGCUCUUGGCAUCACUCACUUCAUUGGAUUCGAUUAGAAAAACGAAUUCAAUGAGUUGCAAGCGAUGCCGCAAUAAAAACUAAUUUGGCAUUGUA-AAA-AGAACAG ......((((((((((....((((((.((((.(((((((((((((.((....)).))))))))))))).))..)).))))))))))).....(......))))))...-...-....... ( -31.60) >DroEre_CAF1 11258 119 - 1 CGUAGAGGUGUAUUACUCUUGGCAACACUUACUUUGUUGUAUUCGAUUAGAAAUACGAAUUCAUUAAGUUGCAAGUGAUGCCGCAAUUAAAACUAUUUCUACGUUGUCAAAA-AAAAUAG (((((((((((.(((.(..(((((.(((((((((...((.(((((..((....))))))).))..))))...))))).)))))..).))).)).))))))))).........-....... ( -29.20) >DroYak_CAF1 9762 114 - 1 CGAAAAGG---AUUACUCUUGGCAACACUCACUUCAUUGGAUUCGAUUACAAUUACGAAUUCAA--UGUUGCAAGUGUUGCCGCAGUAAAAACUAGUUUU-CGUUGUCAAAAAAAAAUAG (((((((.---.(((((..((((((((((..(..(((((((((((..........)))))))))--))..)..)))))))))).)))))...))...)))-))................. ( -34.10) >consensus CGAAAAGGUGUAUUGCUCUUGGCAACACUCACUUCAUUGGAUUCGAUUAGAAAAACGAAUUCAAUGAGUUGCAAGCGAUGCCGCAAUAAAAACUAAUUUGGCGUUGUA_AAA_AAAAUAG ....((((........))))((((((.((((.(((((((((((((..........))))))))))))).))..)).))))))(((((..(((....)))...)))))............. (-20.80 = -21.32 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:29 2006