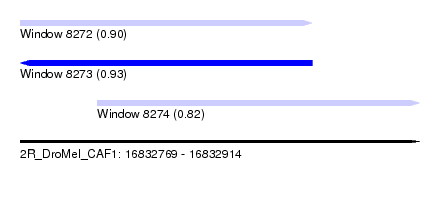
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,832,769 – 16,832,914 |
| Length | 145 |
| Max. P | 0.934727 |

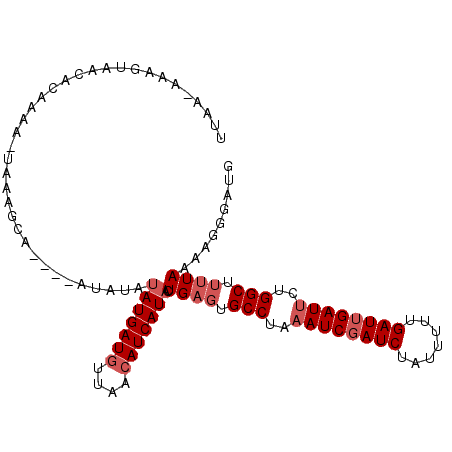
| Location | 16,832,769 – 16,832,875 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -18.10 |
| Energy contribution | -19.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
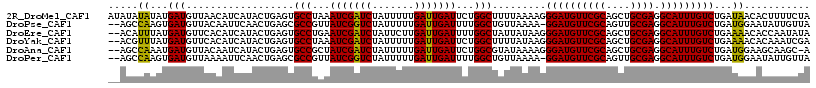
>2R_DroMel_CAF1 16832769 106 + 20766785 UUAAAAAAGUAACACAAAA-UAUAGCCAUAUAUAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGGGAUG ...................-.....((.........(((((((....))))))).((((.(((..((((((((.......))))))))..))).))))...)).... ( -23.70) >DroSec_CAF1 1676 103 + 1 UUUAAAAAGUAACACAAAAAAAAAGCA----AUAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCCAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGGGAUG (((((((....................----.....(((((((....)))))))......(((..((((.(((.......))).))))..))))))))))....... ( -18.70) >DroSim_CAF1 893 102 + 1 UUUAAAAAGUAACACAAAA-AAUAGCA----UUAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGGGAUG ....((((((.........-....(((----((...(((((((....)))))))...)))))(..((((((((.......))))))))..))))))).......... ( -24.00) >DroEre_CAF1 2335 100 + 1 UCG--GAAGUAAUACAAAA-UAACGCA----ACAUUUAUGAUGUUCACAUCAUACUGAGUGCCUGAAUCGAUCUAUUCUUGAUUGAUUUUGGCUAUUAUAAGGGAUG ...--...((((((.....-.......----.....(((((((....)))))))......(((.(((((((((.......))))))))).)))))))))........ ( -23.40) >DroYak_CAF1 903 100 + 1 UUG--AAAGUAAUGCAAAA-UAACACU----ACGUUUAUGAUGUUCACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAUAAGGGAUG .((--(((((.........-...((((----.....(((((((....)))))))...)))).(..((((((((.......))))))))..))))))))......... ( -22.70) >DroAna_CAF1 1243 97 + 1 GCAA-----UAAUGCUUAG-UUAGAUC----AGCCAAAUGAUGUUACAAUCAUACUGAGUGCCGCUAUCGAUCUAUUUUUGAUUGAUUCUGGCGUAUAAAAGGGAUG ((..-----....((((((-(......----......(((((......))))))))))))((((..(((((((.......)))))))..))))))............ ( -22.30) >consensus UUAA_AAAGUAACACAAAA_UAAAGCA____AUAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGGGAUG ....................................(((((((....))))))).((((.(((..((((((((.......))))))))..))).))))......... (-18.10 = -19.10 + 1.00)


| Location | 16,832,769 – 16,832,875 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.85 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -16.35 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16832769 106 - 20766785 CAUCCCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUAUAUAUGGCUAUA-UUUUGUGUUACUUUUUUAA ....((........(((.(((((.(((.......))).))))).))).....((((((((....))))))))........)).....-................... ( -20.10) >DroSec_CAF1 1676 103 - 1 CAUCCCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUGGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUAU----UGCUUUUUUUUUGUGUUACUUUUUAAA .......((((((((((.(((((.(((.......))).))))).))).....((((((((....))))))))....----....................))))))) ( -20.90) >DroSim_CAF1 893 102 - 1 CAUCCCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUAA----UGCUAUU-UUUUGUGUUACUUUUUAAA .......((((((((((.(((((.(((.......))).))))).))).....((((((((....)))))))).(((----(((....-....))))))..))))))) ( -23.00) >DroEre_CAF1 2335 100 - 1 CAUCCCUUAUAAUAGCCAAAAUCAAUCAAGAAUAGAUCGAUUCAGGCACUCAGUAUGAUGUGAACAUCAUAAAUGU----UGCGUUA-UUUUGUAUUACUUC--CGA ..............(((..((((.(((.......))).))))..)))......(((((((....))))))).....----((((...-...)))).......--... ( -18.70) >DroYak_CAF1 903 100 - 1 CAUCCCUUAUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUGAACAUCAUAAACGU----AGUGUUA-UUUUGCAUUACUUU--CAA ..............(((.(((((.(((.......))).))))).)))......(((((((....)))))))...((----(((((..-....)))))))...--... ( -26.60) >DroAna_CAF1 1243 97 - 1 CAUCCCUUUUAUACGCCAGAAUCAAUCAAAAAUAGAUCGAUAGCGGCACUCAGUAUGAUUGUAACAUCAUUUGGCU----GAUCUAA-CUAAGCAUUA-----UUGC ..............(((...(((.(((.......))).)))...)))..((((((((((......)))))...)))----)).....-....(((...-----.))) ( -15.20) >consensus CAUCCCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUAU____UGCUUUA_UUUUGUAUUACUUU_UAAA ..............(((.(((((.(((.......))).))))).))).....((((((((....))))))))................................... (-16.35 = -17.52 + 1.17)



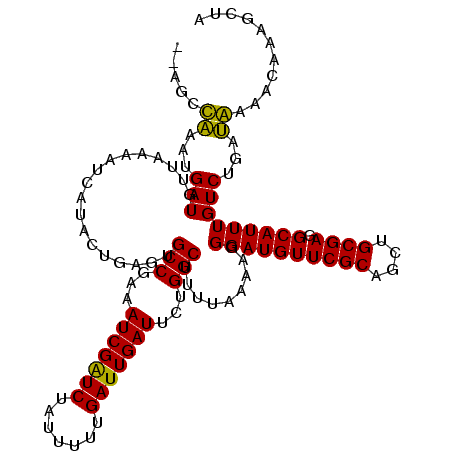
| Location | 16,832,797 – 16,832,914 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -22.30 |
| Energy contribution | -21.13 |
| Covariance contribution | -1.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16832797 117 + 20766785 AUAUAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGGGAUGUUCGCAGCUGCGAGGCAUUUGUCUGAUAACACUUUUCUA .......(((((((....)))))))..(((((((..((((((((.......))))))))..))).........((((((((((....)))).))))))..........))))..... ( -31.20) >DroPse_CAF1 1765 114 + 1 --AGCCAAGUGAUGUUACAAUUCAACUGAGCGCCGUUAUCGGUCUAUUUUUGAUUGAUUUUGGCUGUUAAAA-GGAUGUUCGCAGUUGCGAGGCAUUUGUCUGAUGGAAUAUUGUUA --.((((..(((.........)))..)).))((((..(((((((.......)))))))..))))......((-.(((((((((....)).((((....))))....))))))).)). ( -30.30) >DroEre_CAF1 2359 115 + 1 --ACAUUUAUGAUGUUCACAUCAUACUGAGUGCCUGAAUCGAUCUAUUCUUGAUUGAUUUUGGCUAUUAUAAGGGAUGUUCGCAGCUGCGAGGCAUUUGUCUGAAAACACCAAUAUA --.....(((((((....)))))))((.((((((.(((((((((.......))))))))).))).)))...))((.(((((((....)))((((....))))...))))))...... ( -32.50) >DroYak_CAF1 927 115 + 1 --ACGUUUAUGAUGUUCACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAUAAGGGAUGUUCGCAGCUGCGAGGCAUUUGUCUGAAAACACAAAUCGA --..((((((((((....))))))..((((.(((..((((((((.......))))))))..))).))))....((((((((((....)))).))))))......))))......... ( -31.00) >DroAna_CAF1 1264 114 + 1 --AGCCAAAUGAUGUUACAAUCAUACUGAGUGCCGCUAUCGAUCUAUUUUUGAUUGAUUCUGGCGUAUAAAAGGGAUGUUCGCAGCUGCGAGGCAUUUGUCUGAUGGAAGCAAGC-A --..........((((.(.((((.((.(((((((((((((((((.......)))))))...)))...............((((....))))))))))))).)))).).))))...-. ( -30.70) >DroPer_CAF1 1142 114 + 1 --AGCCAAGUGAUGUUAAAAUUCAACUGAGCGCCGUUAUCGGUCUAUUUUUGAUUGAUUUUGGCUGUUAAAA-GGAUGUUCGCAGUUGCGAGGCAUUUGUCUGAUGGAAUAUUGUUA --.((((..(((.........)))..)).))((((..(((((((.......)))))))..))))......((-.(((((((((....)).((((....))))....))))))).)). ( -30.30) >consensus __AGCCAAAUGAUGUUAAAAUCAUACUGAGUGCCGAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGGGAUGUUCGCAGCUGCGAGGCAUUUGUCUGAUAAAACAAAGCUA .....((...(((..................(((...(((((((.......)))))))...))).........((((((((((....)))).)))))))))...))........... (-22.30 = -21.13 + -1.17)
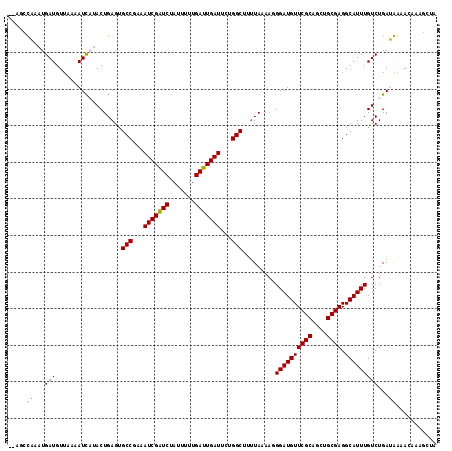
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:24 2006