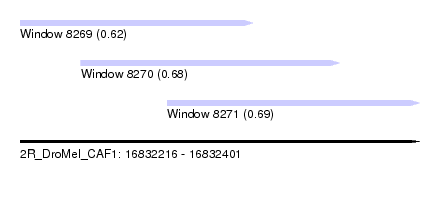
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,832,216 – 16,832,401 |
| Length | 185 |
| Max. P | 0.693859 |

| Location | 16,832,216 – 16,832,324 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -22.54 |
| Energy contribution | -24.02 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16832216 108 + 20766785 ---------GCCAGCAUUGUUGGACCAGCCGAGAGCAGCAACAUUGUAAGCUCCCUGCUACCAGCGUCGGUGGCCUCCAGCAGCGAGGUCGGCGGGCUUUCUUCUACGGCCCUGCAG ---------(((.((..((((((...(((.(.((((.(((....)))..)))).).))).))))))...))(((((((....).)))))))))(((((.........)))))..... ( -39.10) >DroVir_CAF1 81 114 + 1 AGCAAAAACAGCAACAUUGUUGGCGCUGUA---AGCAGCAACAUCGUUGGCUCAUUGCUGCCAGCGUCGGUUGUGGCCAGCAGCGAAGUUGGUGGGGCCAUUUCAACGGCACUGCAG ........((((..(.((((((((((((..---..))))(((..(((((((........)))))))...)))...)))))))).)..))))(..(.(((........))).)..).. ( -45.40) >DroPse_CAF1 1179 105 + 1 ---------AGCAGCAUUGUCGGGACUGCC---AGCAGCAACAUUGUUGGCUCCCUGCUGCCGGCUUCUGUGGCCUCCAGCAGCGAAGUCGGUGGGGUAUCUUCGGAAGCCCUGCAA ---------.((((((.....((((..(((---(((((.....))))))))))))))))))(((((((.((.((.....)).)))))))))(..((((.((....)).))))..).. ( -51.70) >DroSec_CAF1 1120 108 + 1 ---------GCCAGCAUUGUUGGACCAGCUGAGAGCAGCAACAUUGUAAGCUCCCUGCUACCAGCGUCGGUGGCCUCCAGCAGCGAGGUCGGAGGGCUUUCUUCUACGGCCCUGCAG ---------..((((...))))((((.((((.((((.(((....)))..))))...((((((......))))))......))))..))))(.((((((.........)))))).).. ( -38.80) >DroAna_CAF1 698 110 + 1 ----GGAGUAGUAGCAUUGUUGGCACCGCC---AGUAGCAAUAUUGUUGGUUCCUUGCUGCCGGCGUCGGUGGCUGCCAGUAGCGAGGUUGGCGGAGUCUCGUCAACGGCCCUCCAG ----((((.....((...((((((.(((((---(((((((....)))))...(((((((((.(((((.....)).))).))))))))))))))))......)))))).)).)))).. ( -45.30) >DroPer_CAF1 556 105 + 1 ---------AGCAGCAUUGUCGGGACUGCC---AGCAGCAACAUUGUUGGCUCCCUGCUGCCGGCUUCUGUGGCCUCCAGCAGCGAAGUCGGUGGGGUAUCUUCGGAAGCCCUGCAA ---------.((((((.....((((..(((---(((((.....))))))))))))))))))(((((((.((.((.....)).)))))))))(..((((.((....)).))))..).. ( -51.70) >consensus _________AGCAGCAUUGUUGGAACUGCC___AGCAGCAACAUUGUUGGCUCCCUGCUGCCAGCGUCGGUGGCCUCCAGCAGCGAAGUCGGUGGGGUAUCUUCAACGGCCCUGCAG .........(((..(.((((((((...((.....)).((.((..(((((((........)))))))...)).)).)))))))).)..))).((((((((........)))))))).. (-22.54 = -24.02 + 1.48)

| Location | 16,832,244 – 16,832,364 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -45.55 |
| Consensus MFE | -28.69 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16832244 120 + 20766785 GCAACAUUGUAAGCUCCCUGCUACCAGCGUCGGUGGCCUCCAGCAGCGAGGUCGGCGGGCUUUCUUCUACGGCCCUGCAGGACUUGAAUGCCCUCAAGAAGCGCAUACUCCAGCAGAAAU ((......(((.((..(((((..........(.((((((((....).))))))).)(((((.........))))).))))).(((((......)))))....)).)))....))...... ( -37.00) >DroVir_CAF1 115 120 + 1 GCAACAUCGUUGGCUCAUUGCUGCCAGCGUCGGUUGUGGCCAGCAGCGAAGUUGGUGGGGCCAUUUCAACGGCACUGCAGGACUUAAAUGCGCUCAGGAAGCGCGUACUCCAACAGAAAU (((((..(((((((........)))))))...))))).((((((......))))))...(((........))).(((..(((.....(((((((.....)))))))..)))..))).... ( -44.10) >DroGri_CAF1 1280 120 + 1 GCAAUAUUGUUGGCUCAUUGCUGCCGGCGUCAGUUGUGGCCAGCAGCGAGAUUGGUGACGCCGAUUUAACGGCACUGCAAGACUUGAAUGCGCUCAGGAAGCGCAUACUACAACAGAAAU ........((((((........))))))....(((((((...((((.......(....)((((......)))).)))).........(((((((.....))))))).)))))))...... ( -41.60) >DroMoj_CAF1 1281 120 + 1 GCAACAUCGUUGGCUCUUUACUGCCAGCGUCGGUUGUGGCCAGCAGCGAAGUUGGUGGGGCCAUUUCAACGGCACUACAGGACUUGAAUGCGCUCAAGAAGCGCAUUCUCCAACAGAAAU (((((..(((((((........)))))))...))))).((((((......)))))).(((((........))).))...(((...(((((((((.....))))))))))))......... ( -46.90) >DroAna_CAF1 728 120 + 1 GCAAUAUUGUUGGUUCCUUGCUGCCGGCGUCGGUGGCUGCCAGUAGCGAGGUUGGCGGAGUCUCGUCAACGGCCCUCCAGGACCUGAAUGCGCUCAAGAAGCGCAUACUUCAGCAGAAGU ...........(((((((((((((.(((((.....)).))).))))))))((((((((....)))))))).........)))))...(((((((.....)))))))(((((....))))) ( -47.30) >DroPer_CAF1 581 120 + 1 GCAACAUUGUUGGCUCCCUGCUGCCGGCUUCUGUGGCCUCCAGCAGCGAAGUCGGUGGGGUAUCUUCGGAAGCCCUGCAAGACUCGAAUGCGCUCAAGAAGCGCAUUCUCCAGCAGAAAU ......(((((((.((.((((((..((((.....))))..)))))).))((((.(..((((.((....)).))))..)..)))).(((((((((.....))))))))).))))))).... ( -56.40) >consensus GCAACAUUGUUGGCUCCUUGCUGCCAGCGUCGGUGGCGGCCAGCAGCGAAGUUGGUGGGGCCUCUUCAACGGCACUGCAGGACUUGAAUGCGCUCAAGAAGCGCAUACUCCAACAGAAAU ......(((((((.((.((((((.(.(((.....))).).)))))).))((((.((((.(((........))).))))..))))...(((((((.....)))))))...))))))).... (-28.69 = -29.08 + 0.39)

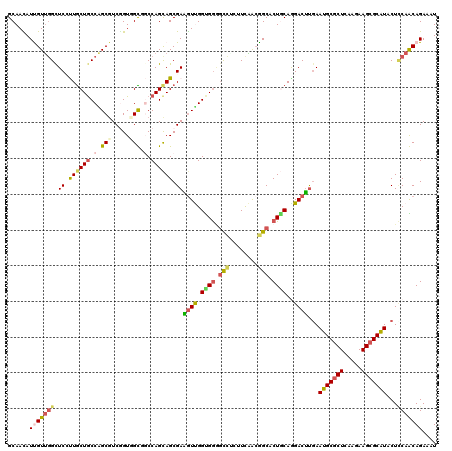
| Location | 16,832,284 – 16,832,401 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -24.99 |
| Energy contribution | -24.05 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
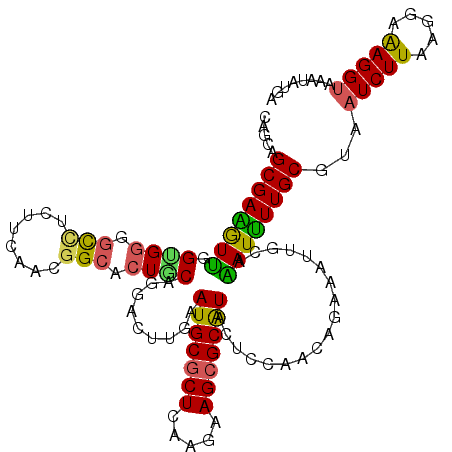
>2R_DroMel_CAF1 16832284 117 + 20766785 CAGCAGCGAGGUCGGCGGGCUUUCUUCUACGGCCCUGCAGGACUUGAAUGCCCUCAAGAAGCGCAUACUCCAGCAGAAAUUGCAGAUCUUGCGUAAUCUUAAAGAAAGGUGAGUUAC- .(((.((((((((.(((((((.........)))))(((.(((...(.((((.((.....)).)))).)))).)))......)).))))))))...(((((.....)))))..)))..- ( -33.50) >DroVir_CAF1 155 118 + 1 CAGCAGCGAAGUUGGUGGGGCCAUUUCAACGGCACUGCAGGACUUAAAUGCGCUCAGGAAGCGCGUACUCCAACAGAAAUAUCAAAUUUUGCGUAAUCUUAAGGAAAGGUAAAUAUGU ..(((((((((((((((..(((........))).(((..(((.....(((((((.....)))))))..)))..)))...)))).))))))))...(((((.....))))).....))) ( -30.00) >DroPse_CAF1 1244 118 + 1 CAGCAGCGAAGUCGGUGGGGUAUCUUCGGAAGCCCUGCAAGACUCGAAUGCGCUCAAGAAGCGCAUUCUCCAGCAGAAAUUGCAGAUUUUGCGUACUCUUAAGGAAAGGUAUGAUUGA .....((((((((.(..((((.((....)).))))..).......(((((((((.....)))))))))....((((...)))).))))))))((((.(((.....)))))))...... ( -43.80) >DroGri_CAF1 1320 118 + 1 CAGCAGCGAGAUUGGUGACGCCGAUUUAACGGCACUGCAAGACUUGAAUGCGCUCAGGAAGCGCAUACUACAACAGAAAUUGCAAAUUUUGCGUAAUCUUAAGGAAAGGUAAAAAUCA .....(((((((((....)((((......))))..(((((.......(((((((.....))))))).((.....))...)))))))))))))...(((((.....)))))........ ( -29.90) >DroMoj_CAF1 1321 118 + 1 CAGCAGCGAAGUUGGUGGGGCCAUUUCAACGGCACUACAGGACUUGAAUGCGCUCAAGAAGCGCAUUCUCCAACAGAAAUAUCAAAUUUUGCGUAAUCUUAAGGAAAGGUAAAUACGA .....((((((((.((((.(((........))).)))).(((...(((((((((.....)))))))))))).............))))))))((((((((.....)))))...))).. ( -35.30) >DroAna_CAF1 768 118 + 1 CAGUAGCGAGGUUGGCGGAGUCUCGUCAACGGCCCUCCAGGACCUGAAUGCGCUCAAGAAGCGCAUACUUCAGCAGAAGUUGCAGAUCUUGCGUAAUCUUAAGGAGAGGUGAGGAAUA ..........((((((((....))))))))...(((((((((.(((.(((((((.....)))))))(((((....)))))..))).)))))....(((((.....))))))))).... ( -37.40) >consensus CAGCAGCGAAGUUGGUGGGGCCUCUUCAACGGCACUGCAGGACUUGAAUGCGCUCAAGAAGCGCAUACUCCAACAGAAAUUGCAAAUUUUGCGUAAUCUUAAGGAAAGGUAAAUAUGA .....((((((((.((((.(((........))).)))).........(((((((.....)))))))..................))))))))...(((((.....)))))........ (-24.99 = -24.05 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:22 2006