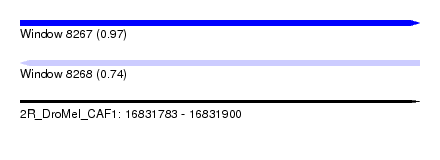
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,831,783 – 16,831,900 |
| Length | 117 |
| Max. P | 0.968333 |

| Location | 16,831,783 – 16,831,900 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -36.85 |
| Energy contribution | -38.43 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
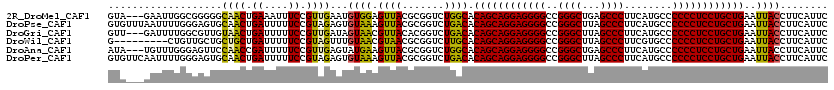
>2R_DroMel_CAF1 16831783 117 + 20766785 GAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGGCCCCUCCUGCUGUGCCAGACCGCGUAACUCCACAUUCAACGGAAAUUUCAGUUGCCCCCGCCAAUUC---UAC ((((...((((...(((((((((((((((.(((.....))))))...)))))))))))))))).......((((((.....(((....))).....))))))........))))---... ( -42.20) >DroPse_CAF1 713 120 + 1 GAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGGCCCCUCCUGCUGUGUCAGACCGCGUAACUUUACACUCUACGGAAAAAUCAGUUGCACUCCCAAAAUUAAACAC .......(((....((((((((..(((((.....((((...)))).)))))))))))))......)))..((((((......((....))......)))))).................. ( -42.70) >DroGri_CAF1 807 117 + 1 GAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGGCCCCUCCUGCUGUGUCAGACCGUGUAACGUUACUAUCAACGGAAAAAUCAGUUACAACGCCAAAAUC---AAC ...(((.(((....((((((((..(((((.....((((...)))).)))))))))))))......))).(((((((((......))).((....)).)))))).........))---).. ( -41.40) >DroWil_CAF1 1374 111 + 1 GAAUGAAGGUAAUUCAGCAGGAGGGGGGCACGAAGGGCUAAGCCCGGCCCCUCCUGCUGUGCAAGACCGCGUUACGUUACAAACUACGGAAAAAUCAGCAGCAGCAACAG---------C .......(((....((((((((..(((((.....((((...)))).)))))))))))))......)))(((((.(((........))).........((....))))).)---------) ( -39.10) >DroAna_CAF1 216 117 + 1 GAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGGCCCCUCCUGCUGUGCCAGACCGCGUAACUUCAUACUCAACGGAAAAAUCGGUUGGAACUCCCAAACA---UAU ..((((((......(((((((((((((((.(((.....))))))...)))))))))))).((......))....))))))..(((((.((....)).)))))............---... ( -43.90) >DroPer_CAF1 81 120 + 1 GAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGGCCCCUCCUGCUGUGUCAGACCGCGUAACUUUACACUCUACGGAAAAAUCAGUUGCACUCCCAAAAUUGAACAC .......(((....((((((((..(((((.....((((...)))).)))))))))))))......)))..((((((......((....))......)))))).................. ( -42.70) >consensus GAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGGCCCCUCCUGCUGUGCCAGACCGCGUAACUUUACACUCAACGGAAAAAUCAGUUGCACCCCCAAAAUC___CAC .......(((....((((((((..(((((.....((((...)))).)))))))))))))......)))..((((((......((....))......)))))).................. (-36.85 = -38.43 + 1.59)

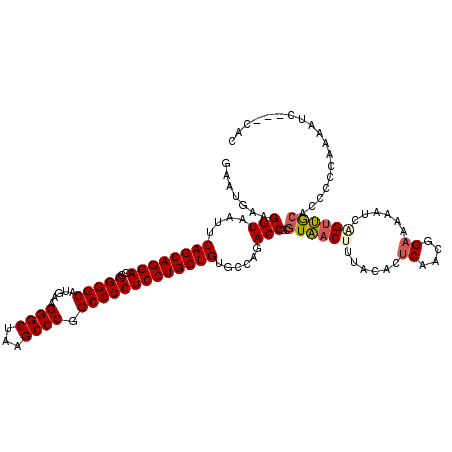
| Location | 16,831,783 – 16,831,900 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -34.28 |
| Energy contribution | -33.45 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16831783 117 - 20766785 GUA---GAAUUGGCGGGGGCAACUGAAAUUUCCGUUGAAUGUGGAGUUACGCGGUCUGGCACAGCAGGAGGGGCCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUC ...---...(..((((..(.........)..))))..)..((((((....((......)).((((((((((((...(((((((...)))).)))))))))))))))......)))))).. ( -46.80) >DroPse_CAF1 713 120 - 1 GUGUUUAAUUUUGGGAGUGCAACUGAUUUUUCCGUAGAGUGUAAAGUUACGCGGUCUGACACAGCAGGAGGGGCCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUC (((((..(((..(((((..(....)...))))).....(((((....))))))))..)))))(((((((((((..((((...))))........)))))))))))............... ( -39.30) >DroGri_CAF1 807 117 - 1 GUU---GAUUUUGGCGUUGUAACUGAUUUUUCCGUUGAUAGUAACGUUACACGGUCUGACACAGCAGGAGGGGCCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUC (((---(((..((((((((((((.((....)).))))....))))))..))..))).))).((((((((((((..((((...))))........)))))))))))).............. ( -40.60) >DroWil_CAF1 1374 111 - 1 G---------CUGUUGCUGCUGCUGAUUUUUCCGUAGUUUGUAACGUAACGCGGUCUUGCACAGCAGGAGGGGCCGGGCUUAGCCCUUCGUGCCCCCCUCCUGCUGAAUUACCUUCAUUC .---------..(((((.(((((.((....)).)))))..)))))((((.(((....))).((((((((((((..((((...))))........))))))))))))..))))........ ( -44.20) >DroAna_CAF1 216 117 - 1 AUA---UGUUUGGGAGUUCCAACCGAUUUUUCCGUUGAGUAUGAAGUUACGCGGUCUGGCACAGCAGGAGGGGCCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUC ...---.............((((.((....)).))))...((((((....((......)).((((((((((((...(((((((...)))).)))))))))))))))......)))))).. ( -42.20) >DroPer_CAF1 81 120 - 1 GUGUUCAAUUUUGGGAGUGCAACUGAUUUUUCCGUAGAGUGUAAAGUUACGCGGUCUGACACAGCAGGAGGGGCCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUC (..(((........)))..).((.((....)).)).(((((((....)))))(((......((((((((((((..((((...))))........))))))))))))....))).)).... ( -39.50) >consensus GUA___AAUUUUGGGGGUGCAACUGAUUUUUCCGUAGAGUGUAAAGUUACGCGGUCUGACACAGCAGGAGGGGCCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUC ...................((((.((....)).))))...((((.((((.......)))).((((((((((((..((((...))))........))))))))))))..))))........ (-34.28 = -33.45 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:19 2006