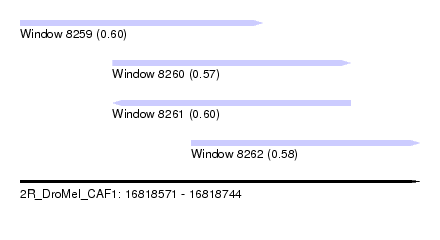
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,818,571 – 16,818,744 |
| Length | 173 |
| Max. P | 0.604778 |

| Location | 16,818,571 – 16,818,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -22.61 |
| Energy contribution | -23.75 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16818571 105 + 20766785 GGACUCAUUAACAGGGAGUUUGCAGAUGGUGUCCACCAUGUGGAAUUCCGGUGGCUCCUCUUUGUUGGCCUUCUGCUGGAUUUGUUCCAGCACCAACUCCGCUCG (((....(((((((((((...((...(((..(((((...)))))...)))...))..))))))))))).....(((((((.....))))))).....)))..... ( -37.70) >DroPse_CAF1 12937 105 + 1 GGACUCGCUCUUAGGCACUUUGUGUAUAGUGUCCACCAUAAGGAACUCCUCCUCCUGAUCCUCCAAUUUUUUCCGCUCGAACAGUUCGAGCACUUGCUUGGCCCG ((.(..((.....((((((........))))))...((..((((.....))))..)).................((((((.....))))))....))..)..)). ( -23.70) >DroSec_CAF1 12669 105 + 1 GGACUCAUUAACAGGGAGCUUGCAGAUGGUGUCCACCAUGUGGAAUUCCCGUGGCUCAUCUUCGUUGGCCUUCUGCUGGAUCUGUUCCAGCACCAACUCCGCACG (((........(((((((.((.((.(((((....))))).)).))))))).))..........(((((.....(((((((.....)))))))))))))))..... ( -37.40) >DroSim_CAF1 12383 105 + 1 GGACUCAUUAACAGGGAGCUUGCAGAUGGUGUCCACCAUGUGGAAUUCCCGUGGCUCAUCUUUGUUGGCCUUCUGCUGGAUCUGUUCCAGCACCAACUCCGCACG (((........(((((((.((.((.(((((....))))).)).))))))).))..........(((((.....(((((((.....)))))))))))))))..... ( -37.10) >DroEre_CAF1 12633 105 + 1 CGACUCAUUAACAGGAAGCUUGCAGAUGGUGUCCACCAUGUGGAAUUCGGGCGGCUCCUCUUUGUUGGCCAUCUGCUGGAUCUGUUCCAGCACCAACUCCGCCCG ..............(((..((.((.(((((....))))).)).)))))((((((.........(((((.....(((((((.....)))))))))))).)))))). ( -37.70) >DroYak_CAF1 12613 105 + 1 CGACUCAUUAACAGGAAGCUUGCAAAUGGCGUCCACCAUAUGGAAUUCGGACGGCUCUUCUUUGUUGGCCAUCUGCUGGAUCUGUUCCAGCACCAAUUCCGCCCG .............(((.(((.......))).))).......((((((.((..((((..........))))...(((((((.....)))))))))))))))..... ( -31.10) >consensus GGACUCAUUAACAGGGAGCUUGCAGAUGGUGUCCACCAUGUGGAAUUCCGGCGGCUCAUCUUUGUUGGCCUUCUGCUGGAUCUGUUCCAGCACCAACUCCGCCCG (((((((((..((((...))))..))))).)))).....(((((........((((..........))))....((((((.....))))))......)))))... (-22.61 = -23.75 + 1.14)

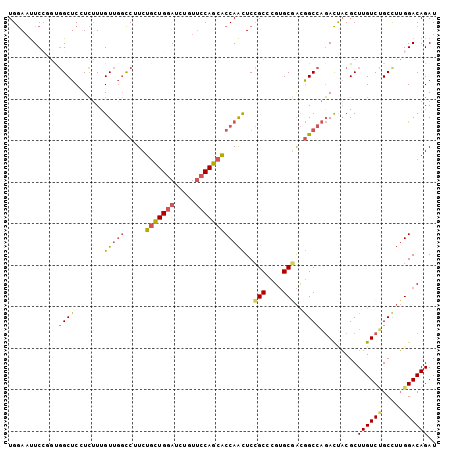
| Location | 16,818,611 – 16,818,714 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.32 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16818611 103 + 20766785 UGGAAUUCCGGUGGCUCCUCUUUGUUGGCCUUCUGCUGGAUUUGUUCCAGCACCAACUCCGCUCGUGCUACGGCCAGACUACGCUUGUCUGCCUUGGACAGAU ......(((((((((........(((((.....(((((((.....)))))))))))).........)))))(((.((((.......))))))).))))..... ( -35.63) >DroSec_CAF1 12709 103 + 1 UGGAAUUCCCGUGGCUCAUCUUCGUUGGCCUUCUGCUGGAUCUGUUCCAGCACCAACUCCGCACGUGCGACGGCCAGACUACGCUUGUCUGCCUUGGACAGAU ........(((.(((...........((((...(((((((.....))))))).......(((....)))..))))((((.......))))))).)))...... ( -34.60) >DroSim_CAF1 12423 103 + 1 UGGAAUUCCCGUGGCUCAUCUUUGUUGGCCUUCUGCUGGAUCUGUUCCAGCACCAACUCCGCACGUGCGACGGCCAGACUACGCUUGUCUGCCUUGGACAGAU ........(((.(((...........((((...(((((((.....))))))).......(((....)))..))))((((.......))))))).)))...... ( -34.60) >DroEre_CAF1 12673 103 + 1 UGGAAUUCGGGCGGCUCCUCUUUGUUGGCCAUCUGCUGGAUCUGUUCCAGCACCAACUCCGCCCGUGCGACGGCCAGACCACGCUUGUCCGCUUUGGACAGAU .......(((((((.........(((((.....(((((((.....)))))))))))).))))))).(((..((.....)).)))((((((.....)))))).. ( -39.00) >DroYak_CAF1 12653 103 + 1 UGGAAUUCGGACGGCUCUUCUUUGUUGGCCAUCUGCUGGAUCUGUUCCAGCACCAAUUCCGCCCGUGCGACAGCCAGACCACGCUUGUCUGCCUUGGACAGAU .((((((.((..((((..........))))...(((((((.....)))))))))))))))(.(((.((((((((........)).)))).))..))).).... ( -34.10) >DroAna_CAF1 14413 103 + 1 UGAACCUCCACUUGCUCCUGGUCGUCCUUCUUCUUUUGGAACAUCAGCAAUGCCAGUUCGGCCCGAGCUGUGGCUCUAUUUCGCUUGUCGGCCAGGGACAGAU ...........(((..(((((((((((..........)))(((..(((...(((.....)))..((((....))))......))))))))))))))..))).. ( -31.90) >consensus UGGAAUUCCGGUGGCUCCUCUUUGUUGGCCUUCUGCUGGAUCUGUUCCAGCACCAACUCCGCCCGUGCGACGGCCAGACUACGCUUGUCUGCCUUGGACAGAU ............((((.......(((((.....(((((((.....))))))))))))..(((....)))..)))).........((((((.....)))))).. (-23.06 = -23.32 + 0.26)

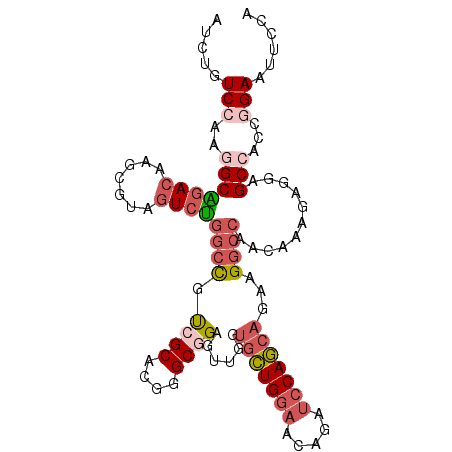
| Location | 16,818,611 – 16,818,714 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -23.36 |
| Energy contribution | -24.92 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16818611 103 - 20766785 AUCUGUCCAAGGCAGACAAGCGUAGUCUGGCCGUAGCACGAGCGGAGUUGGUGCUGGAACAAAUCCAGCAGAAGGCCAACAAAGAGGAGCCACCGGAAUUCCA .(((.(((..(((((((.......)))).)))...((....)))))((((((((((((.....)))))).....))))))..)))(((.((...))...))). ( -34.30) >DroSec_CAF1 12709 103 - 1 AUCUGUCCAAGGCAGACAAGCGUAGUCUGGCCGUCGCACGUGCGGAGUUGGUGCUGGAACAGAUCCAGCAGAAGGCCAACGAAGAUGAGCCACGGGAAUUCCA .(((((.....)))))...........((((((((((....))...((((((((((((.....)))))).....))))))...)))).))))..((....)). ( -35.90) >DroSim_CAF1 12423 103 - 1 AUCUGUCCAAGGCAGACAAGCGUAGUCUGGCCGUCGCACGUGCGGAGUUGGUGCUGGAACAGAUCCAGCAGAAGGCCAACAAAGAUGAGCCACGGGAAUUCCA .(((((.....)))))...........((((((((((....))...((((((((((((.....)))))).....))))))...)))).))))..((....)). ( -35.90) >DroEre_CAF1 12673 103 - 1 AUCUGUCCAAAGCGGACAAGCGUGGUCUGGCCGUCGCACGGGCGGAGUUGGUGCUGGAACAGAUCCAGCAGAUGGCCAACAAAGAGGAGCCGCCCGAAUUCCA ...(((((.....))))).((((((.....))).))).(((((((.((((((((((((.....)))))).....)))))).........)))))))....... ( -43.80) >DroYak_CAF1 12653 103 - 1 AUCUGUCCAAGGCAGACAAGCGUGGUCUGGCUGUCGCACGGGCGGAAUUGGUGCUGGAACAGAUCCAGCAGAUGGCCAACAAAGAAGAGCCGUCCGAAUUCCA ....((((...((.((((.((........))))))))..))))((((((.((((((((.....)))))))(((((((.........).))))))).)))))). ( -40.20) >DroAna_CAF1 14413 103 - 1 AUCUGUCCCUGGCCGACAAGCGAAAUAGAGCCACAGCUCGGGCCGAACUGGCAUUGCUGAUGUUCCAAAAGAAGAAGGACGACCAGGAGCAAGUGGAGGUUCA ...((..(((((.((...(((((....((((....))))..(((.....))).))))).....(((..........))))).)))))..))............ ( -29.00) >consensus AUCUGUCCAAGGCAGACAAGCGUAGUCUGGCCGUCGCACGGGCGGAGUUGGUGCUGGAACAGAUCCAGCAGAAGGCCAACAAAGAGGAGCCACCGGAAUUCCA .....(((..(((((((.......))))((((.((((....))))......(((((((.....)))))))...))))...........)))...)))...... (-23.36 = -24.92 + 1.56)

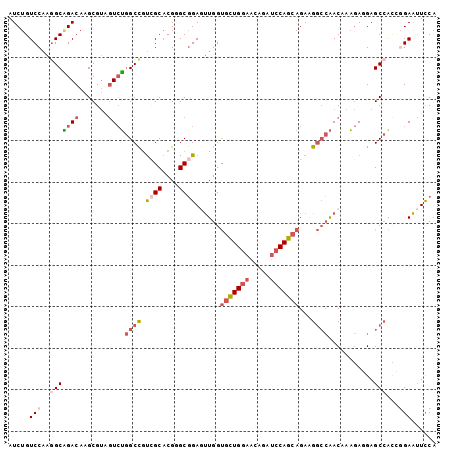
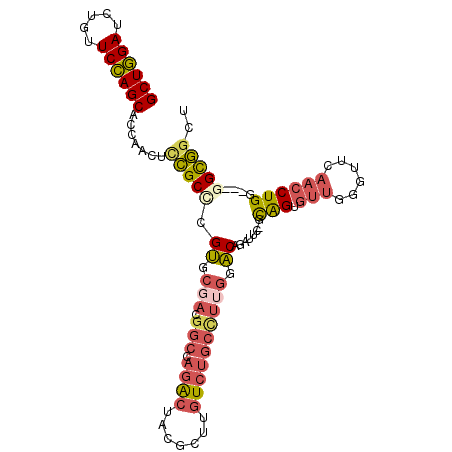
| Location | 16,818,645 – 16,818,744 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -23.78 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16818645 99 + 20766785 GCUGGAUUUGUUCCAGCACCAACUCCGCUCGUGCUACGGCCAGACUACGCUUGUCUGCCUUGGACAGAUUC-ACAGUGUUGAACUCAACCUGG---GGCGGCC ((((((.....)))))).......((((((((.(((.(((.((((.......))))))).)))))......-.(((.((((....))))))))---))))).. ( -40.60) >DroSec_CAF1 12743 99 + 1 GCUGGAUCUGUUCCAGCACCAACUCCGCACGUGCGACGGCCAGACUACGCUUGUCUGCCUUGGACAGAUUC-GCAGUGUUGGGUUCAACCUGG---GGCGGCU ((((((.....)))))).......((((..((.(((.(((.((((.......)))))))))).)).....(-.(((.((((....))))))).---))))).. ( -38.80) >DroSim_CAF1 12457 99 + 1 GCUGGAUCUGUUCCAGCACCAACUCCGCACGUGCGACGGCCAGACUACGCUUGUCUGCCUUGGACAGAUUC-GCAGUGUUGGGUUCAACCUGG---GGCGGCU ((((((.....)))))).......((((..((.(((.(((.((((.......)))))))))).)).....(-.(((.((((....))))))).---))))).. ( -38.80) >DroEre_CAF1 12707 99 + 1 GCUGGAUCUGUUCCAGCACCAACUCCGCCCGUGCGACGGCCAGACCACGCUUGUCCGCUUUGGACAGAUUC-ACAGUGUUAGGUUCAACCUGC---GGUGGCU ((((((.....)))))).(((...(((((((.....)))...((((((((((((((.....))))).....-..)))))..))))......))---))))).. ( -32.61) >DroYak_CAF1 12687 99 + 1 GCUGGAUCUGUUCCAGCACCAAUUCCGCCCGUGCGACAGCCAGACCACGCUUGUCUGCCUUGGACAGAUUU-CCAGUGUUAGGUUCAACCUGC---GGUGUCU ((((((.....)))))).........((....))((((.((((((((((((.((((((....).)))))..-..)))))..))).....))).---).)))). ( -30.80) >DroPer_CAF1 13012 98 + 1 GCUCGAACAGUUCGAGCACUUGCUUGGCCCGACUCUCGGCUAAGUUGCCCUUUUCUCGUUUUGUCAGCUUUUGUGGUGAUGGCU-----CUGGAUUGGCCGCC ((((((.....))))))....((((((((........)))))))).(((....(((.(..(..(((.(......).)))..)..-----).)))..))).... ( -31.20) >consensus GCUGGAUCUGUUCCAGCACCAACUCCGCCCGUGCGACGGCCAGACUACGCUUGUCUGCCUUGGACAGAUUC_GCAGUGUUGGGUUCAACCUGG___GGCGGCU ((((((.....)))))).......(((((.((.(((.(((.((((.......)))))))))).))........(((.(((......))))))....))))).. (-23.78 = -24.62 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:13 2006