
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,800,850 – 16,800,956 |
| Length | 106 |
| Max. P | 0.531808 |

| Location | 16,800,850 – 16,800,956 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.05 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16800850 106 + 20766785 -UAAAUGCCAUAUAAAUAUAGUAUAUAUCGACAUUUAUUGUUAACAUCU----UUACCUGAAUCUGUUGGGCGGCACUCUCCGCAGUCAGUGGCAUGUUGUUUCGAUAUUA -...(((((((....(((((...))))).(((........((((((..(----((....)))..))))))((((......)))).))).)))))))............... ( -20.20) >DroSec_CAF1 7507 105 + 1 -AAAAUGCCAUAAAAAUAUAACAU-UAUCCAUAUUUAUUGUUAACAUCU----UUACCUGAAUCUGUUGGGCGGCACUCUCCGCAGUCAGUGGCAUGUUAUUUCGUUAUUA -...((((((........(((((.-((........)).)))))......----..........(((....((((......))))...)))))))))............... ( -19.10) >DroSim_CAF1 8312 106 + 1 -UAAAUGCCAUAAAAAUAUAACAUAUAUCCAUAUUUAUUGUUAACAUCU----UUACCUGAAUCUGUUGGGCGGCACUCUCCGCAGUUAGUGGCAUUUUUUUUCGUUAUUU -.(((((((((.......((((..................((((((..(----((....)))..))))))((((......)))).)))))))))))))............. ( -19.11) >DroEre_CAF1 7521 97 + 1 -UAAAUGCCAUGGACUUGGAGAAUAAAU-CAUAAGUAUCGUUCACAUUU----UCACCUGAAUCUGUUGGGCGGCACUCUCCGCAGUCAGUGGCAUGUUGUUU-------- -...(((((((.((((((((((......-.....((..((((((((..(----((....)))..)).))))))..)))))))).)))).))))))).......-------- ( -33.50) >DroYak_CAF1 7574 106 + 1 -UAAAUGCCAUGCACUUGGAGCAUAUAUCCAUAAGUAUCGUUCACCUUU----UUACCUGAAUCUGUUGAGCGGCACUUUCCGCAGUCAAUGGCAUUUUGUUUAUUGUUUG -.(((((((((..(((((((............((((..((((((.(..(----((....)))...).))))))..)))))))).)))..)))))))))............. ( -25.00) >DroPer_CAF1 7722 96 + 1 GGAGAUGC-AUCAAGU--UA----AUUUUAUUACUUUCUGUCAUUAUCUUGGCUUACCUGAAUCUGCUGAGCUGCACUCUCUGCAGUUAGUGGCAUUUUGUUA-------- .(((((((-...((((--((----(....((.((.....)).))....)))))))..........(((((.(((((.....)))))))))).)))))))....-------- ( -20.90) >consensus _UAAAUGCCAUAAAAAUAUAGCAUAUAUCCAUAUUUAUUGUUAACAUCU____UUACCUGAAUCUGUUGGGCGGCACUCUCCGCAGUCAGUGGCAUGUUGUUUCGUUAUU_ ....((((((...(((((.............))))).....................((((..((((.(((.......))).))))))))))))))............... (-12.74 = -12.05 + -0.69)

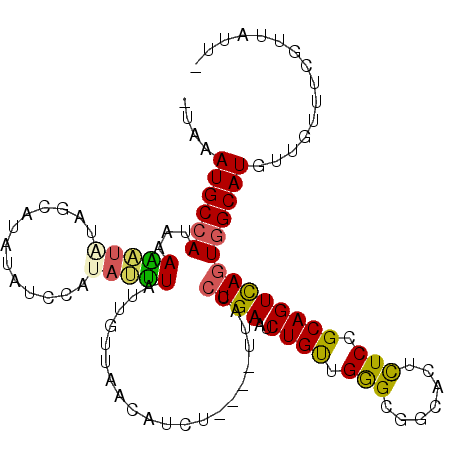
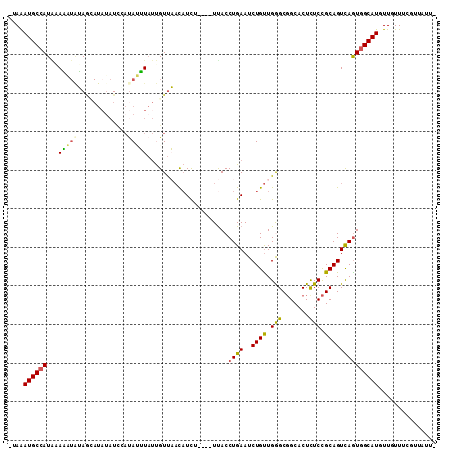
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:05 2006