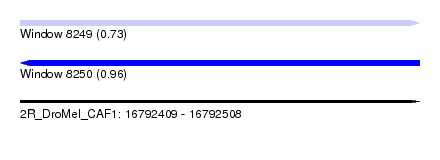
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,792,409 – 16,792,508 |
| Length | 99 |
| Max. P | 0.956648 |

| Location | 16,792,409 – 16,792,508 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.94 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -15.96 |
| Energy contribution | -14.68 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
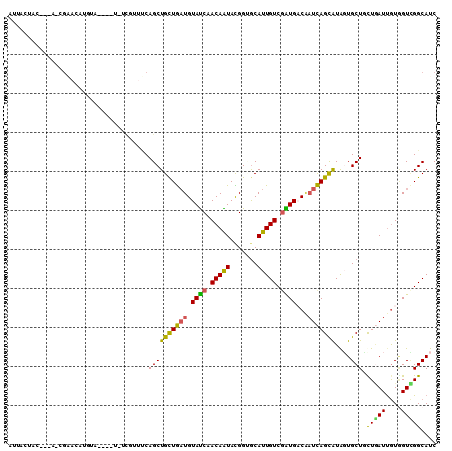
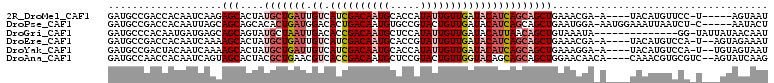
>2R_DroMel_CAF1 16792409 99 + 20766785 AUUACU-----A-GGAACAUGUA----U-UCGUUUCAGCUGCUGAUGUAUCAACAAUAUGGUGCAUUGUCGAUGACAAUCAGCAUAGUGCUCUUGAUUGUGGUCGGCAUC ......-----.-(((((.....----.-..))))).......(((((((((......)))))))))((((((.(((((((((.....))...))))))).))))))... ( -31.00) >DroPse_CAF1 101203 103 + 1 AGUAUU-----G-AGAUUAAUUUCCAUU-UCCAUUCAGCUGCUGAUGUAUCAACAGUACGGCACAUUGUCAGUGUCCAUCAGUGUGCUGCUGCUAAUUGUGGUCGGCAUC ......-----(-((((........)))-))........(((((((....(((.((((((((((((((...........)))))))))).))))..)))..))))))).. ( -26.20) >DroGri_CAF1 141020 95 + 1 AUUGUUAUAAUA-CC--------------UAUUUACAGCUGUUAAUGUAUCAACAAUAUGGAGCAUUGUCGGUGUCAAUUAGCAUACUGCUGCUCAUCAUUGUGGGCAUC ............-..--------------........((((.((((((.(((......))).)))))).)))).......(((.....)))((((((....))))))... ( -18.50) >DroEre_CAF1 102042 102 + 1 AUUUCUACU--A-UGGACAUGUA----U-UCGUUUCAGCUGCUGAUGUAUCAACAAUACGGUGCAUUGUCGAUGACAAUCAGCAUAGUGCUUUUGAUUGUGGUCGGCAUC .........--(-((.((.((((----(-(.(((((((...))))......))))))))))).)))(((((((.(((((((((.....))...))))))).))))))).. ( -28.00) >DroYak_CAF1 108536 102 + 1 AUUACUACA--A-UGGACAUGUA----U-UCCUUUCAGCUGCUGAUGUAUCAACAAUAUGGUGCAUUGUCGAUGACAAUCAGCAUAGUGCUUUUGAUUGUAGUCGGCAUC .........--.-.(((......----.-)))...........(((((((((......)))))))))((((((.(((((((((.....))...))))))).))))))... ( -26.40) >DroAna_CAF1 126861 104 + 1 CUUGAUACU--GACGCACGUUUG----UGUUGUUCCAGCUGCUGCUGUACCAACAGUACGGAGCAUUGUCGGUGACGUUCAGCGUAGUGCUACUGAUUGUGGUUGGCAUC ...(((((.--(((....))).)----))))...(((((..(((((((....))))))((((((((((.((.((.....)).))))))))).)))...)..))))).... ( -35.00) >consensus AUUACUAC___A_CGAACAUGUA____U_UCGUUUCAGCUGCUGAUGUAUCAACAAUACGGUGCAUUGUCGAUGACAAUCAGCAUAGUGCUGCUGAUUGUGGUCGGCAUC ....................................((((((((((.((((.(((((.......))))).))))...)))))))....))).(((((....))))).... (-15.96 = -14.68 + -1.27)

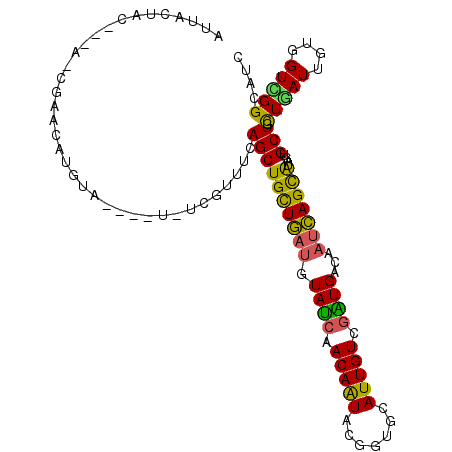
| Location | 16,792,409 – 16,792,508 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.94 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -13.75 |
| Energy contribution | -15.17 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16792409 99 - 20766785 GAUGCCGACCACAAUCAAGAGCACUAUGCUGAUUGUCAUCGACAAUGCACCAUAUUGUUGAUACAUCAGCAGCUGAAACGA-A----UACAUGUUCC-U-----AGUAAU ..((((............).)))...(((((((.((.((((((((((.....)))))))))))))))))))((((.((((.-.----....))))..-)-----)))... ( -22.40) >DroPse_CAF1 101203 103 - 1 GAUGCCGACCACAAUUAGCAGCAGCACACUGAUGGACACUGACAAUGUGCCGUACUGUUGAUACAUCAGCAGCUGAAUGGA-AAUGGAAAUUAAUCU-C-----AAUACU (((.(((....((.(((((.((.(((((.((..(....)...)).))))).))..((((((....))))))))))).))..-..)))......))).-.-----...... ( -22.60) >DroGri_CAF1 141020 95 - 1 GAUGCCCACAAUGAUGAGCAGCAGUAUGCUAAUUGACACCGACAAUGCUCCAUAUUGUUGAUACAUUAACAGCUGUAAAUA--------------GG-UAUUAUAACAAU ((((((.....(((((..((((((((((...((((.......))))....))))))))))...)))))((....)).....--------------))-))))........ ( -20.40) >DroEre_CAF1 102042 102 - 1 GAUGCCGACCACAAUCAAAAGCACUAUGCUGAUUGUCAUCGACAAUGCACCGUAUUGUUGAUACAUCAGCAGCUGAAACGA-A----UACAUGUCCA-U--AGUAGAAAU ......(((............((...(((((((.((.(((((((((((...))))))))))))))))))))..))......-.----.....)))..-.--......... ( -24.35) >DroYak_CAF1 108536 102 - 1 GAUGCCGACUACAAUCAAAAGCACUAUGCUGAUUGUCAUCGACAAUGCACCAUAUUGUUGAUACAUCAGCAGCUGAAAGGA-A----UACAUGUCCA-U--UGUAGUAAU .......((((((((....(((....(((((((.((.((((((((((.....))))))))))))))))))))))....(((-.----......))))-)--))))))... ( -30.90) >DroAna_CAF1 126861 104 - 1 GAUGCCAACCACAAUCAGUAGCACUACGCUGAACGUCACCGACAAUGCUCCGUACUGUUGGUACAGCAGCAGCUGGAACAACA----CAAACGUGCGUC--AGUAUCAAG (((((..(((((..(((((........)))))...(((((((((.(((...))).))))))).((((....))))))......----.....))).)).--.)))))... ( -29.40) >consensus GAUGCCGACCACAAUCAAAAGCACUAUGCUGAUUGUCACCGACAAUGCACCAUAUUGUUGAUACAUCAGCAGCUGAAACGA_A____UACAUGUCCA_U___GUAGUAAU ...................(((....(((((((.((.((((((((((.....)))))))))))))))))))))).................................... (-13.75 = -15.17 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:02 2006