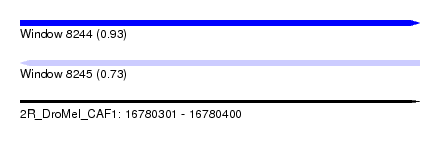
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,780,301 – 16,780,400 |
| Length | 99 |
| Max. P | 0.929369 |

| Location | 16,780,301 – 16,780,400 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.21 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -18.19 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16780301 99 + 20766785 UGUUUUUUUCCU---------CCCUUUAUAUUCUACAUUUGUGUCACGUUAUUGUUACACAUACGCACUGUGUG-CCAUCGCAACCCACUUACAGUGCGAUUCGGAACU .......((((.---------..................(((((.(((....))).)))))..(((((((((((-(....))).......)))))))))....)))).. ( -24.31) >DroPse_CAF1 86783 107 + 1 --UGUUUUUCCUUUUUGCAUGCCUUUUGUAUUCUAUAUUUGUGUCACGUUGUUGUUACACAUACGCACUGUGGCAGGAUAGCGCUCUAUCUACAGGGCCUAACGAAAGU --....................(((((((.........((.(((((((.(((.((.......))))).))))))).))(((.(((((......))))))))))))))). ( -24.50) >DroSec_CAF1 92325 97 + 1 --UUUUUUUCCU---------CCCUUUAUAUUCUACAUUUGUGUCACGUUAUUGUUACACAUACGCACUGUGUG-CCAUCGCAACCCACUUACAGUGCGACUCGGAACU --.....((((.---------..................(((((.(((....))).)))))..(((((((((((-(....))).......)))))))))....)))).. ( -24.31) >DroSim_CAF1 91256 97 + 1 --UUUUUUUCCU---------CCCUAUAUAUUCUACAUUUGUGUCACGUUAUUGUUACACAUACGCACUGUGUG-CCAUCGCAACCCACUUACAGUGCGUCUCAAAACU --..........---------..................(((((.(((....))).))))).((((((((((((-(....))).......))))))))))......... ( -21.01) >DroEre_CAF1 90159 95 + 1 --U-UUUUUCCU---------UCCUUUAUAUUCUACAUUUGUGUCACGUUAUUGUUACACAUACGCACUGUGGG-CCAUCCCAACCA-CUUACAGUGCGACGCGGAACU --.-...((((.---------..................(((((.(((....))).)))))..(((((((((((-............-)))))))))))....)))).. ( -26.10) >DroYak_CAF1 96464 95 + 1 --U-UUUUUCCU---------CCCUUUAUGUUCGACAUUUGUGUCACGUUAUUGUUACACAUACGCACUGUGUG-CCAUCUCAACCC-ACUACAGUGCGAGUCGGAACU --.-........---------.........((((((...(((((.(((....))).)))))..(((((((((((-...........)-)).)))))))).))))))... ( -28.70) >consensus __UUUUUUUCCU_________CCCUUUAUAUUCUACAUUUGUGUCACGUUAUUGUUACACAUACGCACUGUGUG_CCAUCGCAACCCACUUACAGUGCGACUCGGAACU .......((((............................(((((.(((....))).)))))..(((((((((..................)))))))))....)))).. (-18.19 = -19.02 + 0.83)

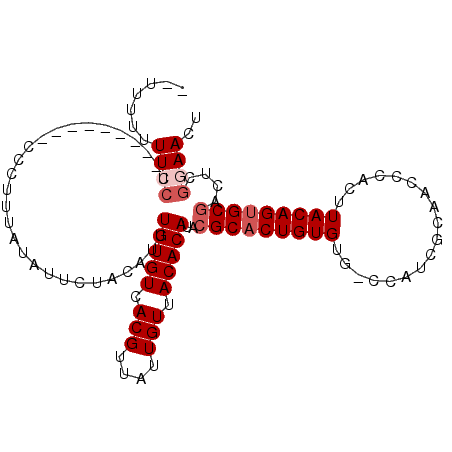
| Location | 16,780,301 – 16,780,400 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.21 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -17.84 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16780301 99 - 20766785 AGUUCCGAAUCGCACUGUAAGUGGGUUGCGAUGG-CACACAGUGCGUAUGUGUAACAAUAACGUGACACAAAUGUAGAAUAUAAAGGG---------AGGAAAAAAACA ..((((....((((((((..((.(.......).)-)..))))))))..(((((.((......)).)))))..................---------.))))....... ( -23.30) >DroPse_CAF1 86783 107 - 1 ACUUUCGUUAGGCCCUGUAGAUAGAGCGCUAUCCUGCCACAGUGCGUAUGUGUAACAACAACGUGACACAAAUAUAGAAUACAAAAGGCAUGCAAAAAGGAAAAACA-- .((((.((...(((.((((......(((((..........)))))...(((((.((......)).))))).........))))...)))..))..))))........-- ( -19.70) >DroSec_CAF1 92325 97 - 1 AGUUCCGAGUCGCACUGUAAGUGGGUUGCGAUGG-CACACAGUGCGUAUGUGUAACAAUAACGUGACACAAAUGUAGAAUAUAAAGGG---------AGGAAAAAAA-- ..((((....((((((((..((.(.......).)-)..))))))))..(((((.((......)).)))))..................---------.)))).....-- ( -23.30) >DroSim_CAF1 91256 97 - 1 AGUUUUGAGACGCACUGUAAGUGGGUUGCGAUGG-CACACAGUGCGUAUGUGUAACAAUAACGUGACACAAAUGUAGAAUAUAUAGGG---------AGGAAAAAAA-- .((((((..(((((((((..((.(.......).)-)..))))))))).(((((.((......)).)))))....))))))........---------..........-- ( -22.60) >DroEre_CAF1 90159 95 - 1 AGUUCCGCGUCGCACUGUAAG-UGGUUGGGAUGG-CCCACAGUGCGUAUGUGUAACAAUAACGUGACACAAAUGUAGAAUAUAAAGGA---------AGGAAAAA-A-- .((((..(((((((((((..(-.((((.....))-)))))))))))..(((((.((......)).))))).)))..))))........---------........-.-- ( -25.80) >DroYak_CAF1 96464 95 - 1 AGUUCCGACUCGCACUGUAGU-GGGUUGAGAUGG-CACACAGUGCGUAUGUGUAACAAUAACGUGACACAAAUGUCGAACAUAAAGGG---------AGGAAAAA-A-- .((((.(((.((((((((.((-(...........-)))))))))))..(((((.((......)).)))))...)))))))........---------........-.-- ( -27.60) >consensus AGUUCCGAGUCGCACUGUAAGUGGGUUGCGAUGG_CACACAGUGCGUAUGUGUAACAAUAACGUGACACAAAUGUAGAAUAUAAAGGG_________AGGAAAAAAA__ ..((((....((((((((....................))))))))..(((((.((......)).)))))............................))))....... (-17.84 = -17.82 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:57 2006