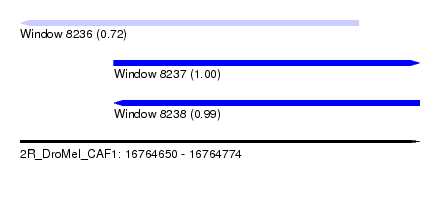
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,764,650 – 16,764,774 |
| Length | 124 |
| Max. P | 0.997862 |

| Location | 16,764,650 – 16,764,755 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.34 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -2.14 |
| Energy contribution | -1.74 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.12 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.719013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16764650 105 - 20766785 GAGCUGCAGAAACAUGAAACUGCAUACGAG----UAAAGUUGCAUGUAUAUGAAAAACAAACAUGCCACAUAAUUCCAGGAAAAUGUAAUUACU-GUAACCAAAAGUUCA (((((((((..........)))).(((.((----(((.((.((((((.............)))))).)).......((......))...)))))-)))......))))). ( -20.32) >DroSec_CAF1 76797 84 - 1 GAGCUGCACAAA--------------CGAG----UAUAUUUGCA------UGAAAAACAAACAUGCAACAUAAUUCCAGGGAAAUGUAAUUACU-GUGACCG-AAGUUCU (((((......(--------------(.((----((....((((------((.........))))))((((..(((....)))))))...))))-)).....-.))))). ( -17.40) >DroSim_CAF1 75004 98 - 1 GAGCUGCACAAACAUGAAACAGCACACGAG----UAUAUUUGCA------UGAAAAACAAACAUGCAACAUAAUUCCAGGGAAAUGUAAUUACU-GUGACCG-AAGUUCA (((((((..............)).(((.((----((....((((------((.........))))))((((..(((....)))))))...))))-)))....-.))))). ( -19.14) >DroEre_CAF1 74165 98 - 1 GAGUUUCCCAAACAUGAAACUGCAUACUAUACGGGU---AUGUA------UGA-AAAGAAACAUGCAACAUAAUUCCUGGAAAAUGUAAUUGCU-AUGAGCA-AAGUUCU .((((((........))))))(((((((.....)))---)))).------...-..((((((((....((.......))....))))..((((.-....)))-)..)))) ( -16.80) >DroYak_CAF1 80147 96 - 1 GAGUUUCACAAACGUGAAACUGCAUAUAGU----UA---AUACA------UGAGAAACAAACAUGCAACAUAAUUCUUGAGAAAUGUAAUUACUAAUGACCA-AAGUUCA .((((((((....)))))))).(((.((((----..---.((((------((((((.................))))).....)))))...)))))))....-....... ( -15.43) >consensus GAGCUGCACAAACAUGAAACUGCAUACGAG____UA_A_UUGCA______UGAAAAACAAACAUGCAACAUAAUUCCAGGGAAAUGUAAUUACU_GUGACCA_AAGUUCA (((((...................................((((......(....).......))))((((..((....))..)))).................))))). ( -2.14 = -1.74 + -0.40)


| Location | 16,764,679 – 16,764,774 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.71 |
| Mean single sequence MFE | -19.23 |
| Consensus MFE | -10.49 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16764679 95 + 20766785 CCUGGAAUUAUGUGGCAUGUUUGUUUUUCAUAUACAUGCAACUUUA----CUCGUAUGCAGUUUCAUGUUUCUGCAGCUCAUCAUUCCGUUCUUUUUUU ...(((((.(((.(((((((.(((.....))).)))))).......----......(((((..........))))).).))).)))))........... ( -21.40) >DroEre_CAF1 74193 85 + 1 CCAGGAAUUAUGUUGCAUGUUUCUUU-UCA------UACAU---ACCCGUAUAGUAUGCAGUUUCAUGUUUGGGAAACUCAUCAUUCCGUUUUUG---- ...(((((.(((..(((((...((..-.((------(((((---(....))).))))).))...)))))..((....))))).))))).......---- ( -17.90) >DroYak_CAF1 80176 82 + 1 UCAAGAAUUAUGUUGCAUGUUUGUUUCUCA------UGUAU---UA----ACUAUAUGCAGUUUCACGUUUGUGAAACUCAUCAUUCGGUUUUUG---- ....((((.(((.(((((((..(((.....------.....---.)----)).)))))))(((((((....))))))).))).))))........---- ( -18.40) >consensus CCAGGAAUUAUGUUGCAUGUUUGUUU_UCA______UGCAU___UA____AUAGUAUGCAGUUUCAUGUUUGUGAAACUCAUCAUUCCGUUUUUG____ ...(((((.(((..((((((.................................))))))((((((((....))))))))))).)))))........... (-10.49 = -11.28 + 0.78)


| Location | 16,764,679 – 16,764,774 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.71 |
| Mean single sequence MFE | -17.47 |
| Consensus MFE | -8.99 |
| Energy contribution | -10.32 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16764679 95 - 20766785 AAAAAAAGAACGGAAUGAUGAGCUGCAGAAACAUGAAACUGCAUACGAG----UAAAGUUGCAUGUAUAUGAAAAACAAACAUGCCACAUAAUUCCAGG ...........(((((.(((...(((((..........)))))......----.......((((((.............))))))..))).)))))... ( -18.12) >DroEre_CAF1 74193 85 - 1 ----CAAAAACGGAAUGAUGAGUUUCCCAAACAUGAAACUGCAUACUAUACGGGU---AUGUA------UGA-AAAGAAACAUGCAACAUAAUUCCUGG ----.......(((((.(((((((((........))))))(((((((.....)))---)))).------...-..............))).)))))... ( -18.70) >DroYak_CAF1 80176 82 - 1 ----CAAAAACCGAAUGAUGAGUUUCACAAACGUGAAACUGCAUAUAGU----UA---AUACA------UGAGAAACAAACAUGCAACAUAAUUCUUGA ----........((((.(((((((((((....))))))))((((...((----(.---.....------.....)))....))))..))).)))).... ( -15.60) >consensus ____CAAAAACGGAAUGAUGAGUUUCACAAACAUGAAACUGCAUACAAU____UA___AUGCA______UGA_AAACAAACAUGCAACAUAAUUCCUGG ...........(((((.((((((((((......)))))))((((.....................................))))..))).)))))... ( -8.99 = -10.32 + 1.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:50 2006