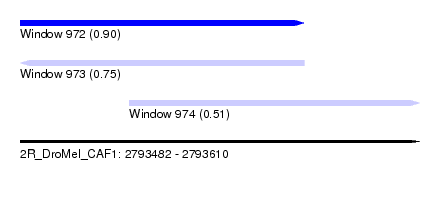
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,793,482 – 2,793,610 |
| Length | 128 |
| Max. P | 0.904267 |

| Location | 2,793,482 – 2,793,573 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -21.46 |
| Consensus MFE | -18.98 |
| Energy contribution | -20.44 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
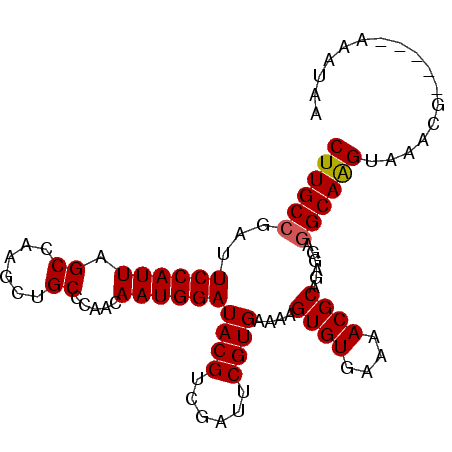
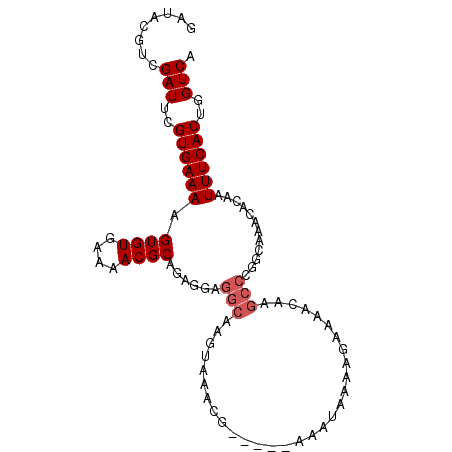
>2R_DroMel_CAF1 2793482 91 + 20766785 CUUGCCGAUUCCAUUAGCCAAGUUGCCCAACAAUGGAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAAGUAAACG-----AAAUAA ((((((..(((......(((.(((....)))..))).((((......))))....((((....)))).)))..))))))......-----...... ( -21.40) >DroSec_CAF1 381 91 + 1 CUUGCCGAUUCCAUUAGCCAAGCUGCCCAACAAUGGAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGACGCAGGUAAACG-----AAAUAA ((((((((((((((((((...)))).......)))))...))))...........((((....)))).......)))))......-----...... ( -18.11) >DroSim_CAF1 386 91 + 1 CUUGCCGAUUCCAUUAGCCAAGCUGCCCAACAAUGGAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAGGUAAACG-----AAAUAA ((((((...(((((((((...)))).......)))))((((......))))....((((....))))......))))))......-----...... ( -21.01) >DroEre_CAF1 387 96 + 1 CUUGCCGAUUCCAUUAGCAAAGCUGCCCAACAAUGGAUACGUUGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAAGAAAAUGAAAUGAAAUAA ((((((..((((((..(((....)))....(((((....)))))....)))....((((....)))).)))..))))))................. ( -23.60) >DroYak_CAF1 393 91 + 1 CUUGCCGAUUCCAUUAGCAAAGCUGCCCAACAAUGGAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAAAAGAGGCAAGAAAAUG-----AAAUAA ((((((...((((((.(((....))).....))))))((((......))))....((((....))))......))))))......-----...... ( -23.20) >consensus CUUGCCGAUUCCAUUAGCCAAGCUGCCCAACAAUGGAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAAGUAAACG_____AAAUAA ((((((...((((((.((......)).....))))))((((......))))....((((....))))......))))))................. (-18.98 = -20.44 + 1.46)

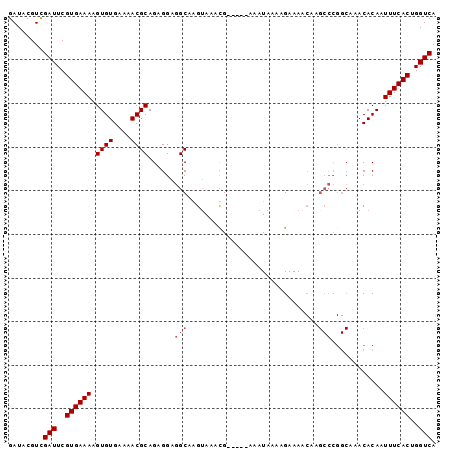
| Location | 2,793,482 – 2,793,573 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
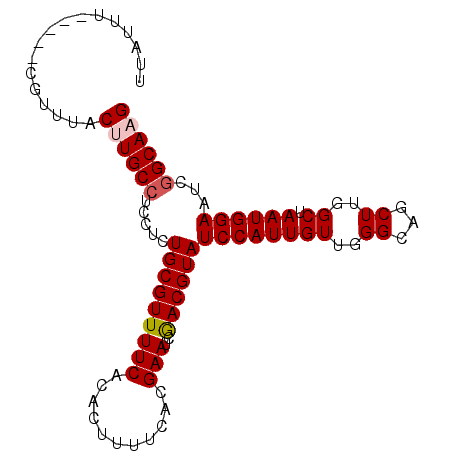
>2R_DroMel_CAF1 2793482 91 - 20766785 UUAUUU-----CGUUUACUUGCCUCCUCUGCGUUUUCACACUUUUCACGAAUCGACGUAUCCAUUGUUGGGCAACUUGGCUAAUGGAAUCGGCAAG ......-----......((((((.....(((((((((...........)))..))))))((((((((..((...))..)).))))))...)))))) ( -23.70) >DroSec_CAF1 381 91 - 1 UUAUUU-----CGUUUACCUGCGUCCUCUGCGUUUUCACACUUUUCACGAAUCGACGUAUCCAUUGUUGGGCAGCUUGGCUAAUGGAAUCGGCAAG ......-----......((((((((.....(((.............)))....))))))((((((((..((...))..)).))))))...)).... ( -18.42) >DroSim_CAF1 386 91 - 1 UUAUUU-----CGUUUACCUGCCUCCUCUGCGUUUUCACACUUUUCACGAAUCGACGUAUCCAUUGUUGGGCAGCUUGGCUAAUGGAAUCGGCAAG ......-----........((((.....(((((((((...........)))..))))))((((((((..((...))..)).))))))...)))).. ( -20.60) >DroEre_CAF1 387 96 - 1 UUAUUUCAUUUCAUUUUCUUGCCUCCUCUGCGUUUUCACACUUUUCACGAAUCAACGUAUCCAUUGUUGGGCAGCUUUGCUAAUGGAAUCGGCAAG .................((((((.....(((((((((...........)))..))))))((((((....(((......)))))))))...)))))) ( -20.10) >DroYak_CAF1 393 91 - 1 UUAUUU-----CAUUUUCUUGCCUCUUUUGCGUUUUCACACUUUUCACGAAUCGACGUAUCCAUUGUUGGGCAGCUUUGCUAAUGGAAUCGGCAAG ......-----......((((((.....(((((((((...........)))..))))))((((((....(((......)))))))))...)))))) ( -20.60) >consensus UUAUUU_____CGUUUACUUGCCUCCUCUGCGUUUUCACACUUUUCACGAAUCGACGUAUCCAUUGUUGGGCAGCUUGGCUAAUGGAAUCGGCAAG .................((((((.....(((((((((...........)))..))))))((((((((..((...))..)).))))))...)))))) (-19.26 = -19.70 + 0.44)

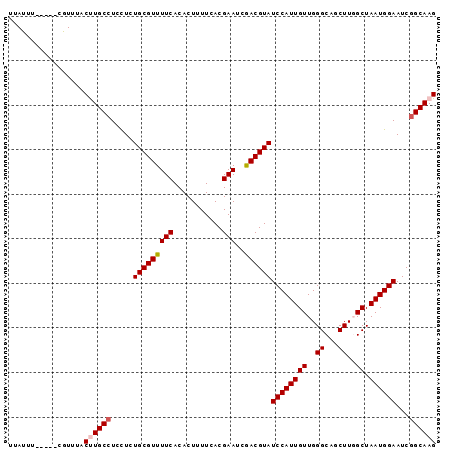
| Location | 2,793,517 – 2,793,610 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -17.22 |
| Consensus MFE | -13.05 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2793517 93 + 20766785 GAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAAGUAAACG-----AAAUAAAAGAAAACAAGCCCGGCAAACACAAUUUCACUGGUCA ........(((..((((((.((((....)))).(..(.(((..((.....-----............))..))))..)........))))))..))). ( -17.93) >DroSec_CAF1 416 80 + 1 GAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGACGCAGGUAAACG-----AAAUAAAAGAAA-------------ACACAAUUUCACUGGUCA ........(((..((((((.((((....))))......((........))-----............-------------......))))))..))). ( -14.80) >DroSim_CAF1 421 93 + 1 GAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAGGUAAACG-----AAAUAAAAGAAAACAAGCCCGGCAAACACAAUUUCACUGGUCA ........(((..((((((.((((....)))).(..(.(((..((.....-----............))..))))..)........))))))..))). ( -18.93) >DroEre_CAF1 422 98 + 1 GAUACGUUGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAAGAAAAUGAAAUGAAAUAAAAGAAAACAAGCCCGGCAAACACAAUUUCACUGGUCA .......((((..((((((.((((....)))).(..(.(((..............................))))..)........))))))..)))) ( -17.21) >consensus GAUACGUCGAUUCGUGAAAAGUGUGAAAACGCAGAGGAGGCAAGUAAACG_____AAAUAAAAGAAAACAAGCCCGGCAAACACAAUUUCACUGGUCA ........(((..((((((..((((.....((.......))..............................((...))...)))).))))))..))). (-13.05 = -13.18 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:40 2006