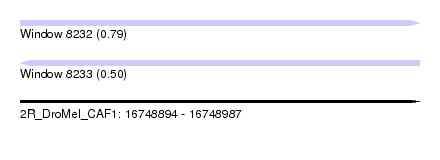
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,748,894 – 16,748,987 |
| Length | 93 |
| Max. P | 0.792856 |

| Location | 16,748,894 – 16,748,987 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -14.65 |
| Consensus MFE | -8.62 |
| Energy contribution | -9.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
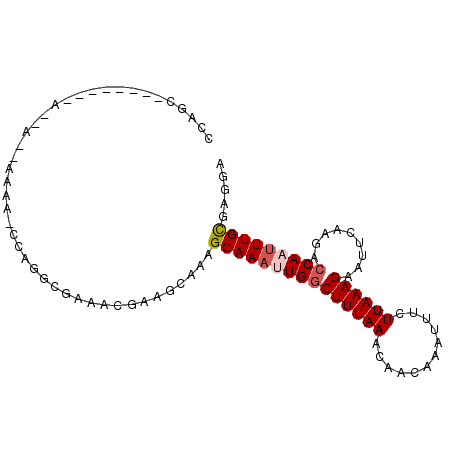
>2R_DroMel_CAF1 16748894 93 + 20766785 CCAGCU------CAGGAAAAAAAACCAGGCGAAACGAAGCAAAGCAAAUUGGCUUUAAACAACAAAUUUCUUAAAGAAUUCAAGACCAAUUUGCGAGAA ....((------(.((........))..((........))...((((((((((((.((.........(((.....))))).))).)))))))))))).. ( -17.20) >DroSec_CAF1 61100 75 + 1 CCAGU------------------------AAAAACGAAGCAUAGCAAAUUGGCUUUAAACAACAAAUUUCUUAAAGAAUUCAAGAACAAUUUGCGAGGA ((.((------------------------....))........((((((((.((((((............))))))..........))))))))..)). ( -10.50) >DroSim_CAF1 58883 75 + 1 CCAGC------------------------AUAAACGAAGCAUAGCAAAUUGGCUUUAAACAACAAAUUUCUUAAAGAAUUCAAGACCAAUUUGCGAGGA ((.((------------------------.........))...((((((((((((.((.........(((.....))))).))).)))))))))..)). ( -14.10) >DroEre_CAF1 58095 98 + 1 CCAGCAAAAAAUAAAUAGGAAAAGCCAGGCGAAACCAACCACAGCAAAUUGGCUUUAAACAACAAAUUUCUUAAAGAAUUCGAGACCAU-UUGUGAGGA ((..................(((((((.((.............))....))))))).....((((((.((((.........))))..))-))))..)). ( -14.52) >DroYak_CAF1 63778 91 + 1 --------AAAUAAAUAGGAAAAGCCAGGCGAAUUGAACCAAAGCAAAUUGGCUUUAAGCAACAAAUUUCUUAAAGAAUUGGAGACCAUUUUGUGAGGA --------............(((((((.((.............))....)))))))...........((((((.(((((.(....).))))).)))))) ( -16.92) >consensus CCAGC________A__A__AAAA_CCAGGCGAAACGAAGCAAAGCAAAUUGGCUUUAAACAACAAAUUUCUUAAAGAAUUCAAGACCAAUUUGCGAGGA ...........................................(((((((((((((((............)))))).........)))))))))..... ( -8.62 = -9.18 + 0.56)


| Location | 16,748,894 – 16,748,987 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -16.03 |
| Consensus MFE | -10.22 |
| Energy contribution | -12.22 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
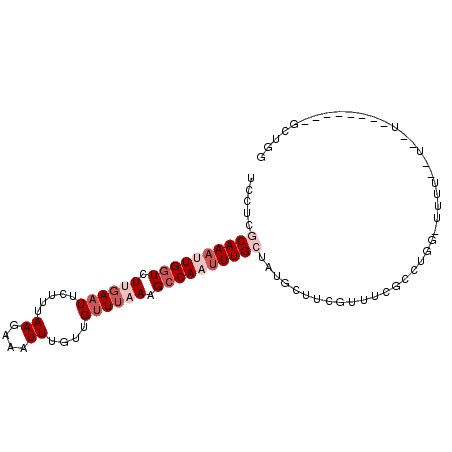
>2R_DroMel_CAF1 16748894 93 - 20766785 UUCUCGCAAAUUGGUCUUGAAUUCUUUAAGAAAUUUGUUGUUUAAAGCCAAUUUGCUUUGCUUCGUUUCGCCUGGUUUUUUUUCCUG------AGCUGG .....(((((((((..((((((.....((....))....))))))..)))))))))...............(..((((........)------)))..) ( -22.50) >DroSec_CAF1 61100 75 - 1 UCCUCGCAAAUUGUUCUUGAAUUCUUUAAGAAAUUUGUUGUUUAAAGCCAAUUUGCUAUGCUUCGUUUUU------------------------ACUGG .....(((((((((((((((.....)))))))....(((......)))))))))))..............------------------------..... ( -12.90) >DroSim_CAF1 58883 75 - 1 UCCUCGCAAAUUGGUCUUGAAUUCUUUAAGAAAUUUGUUGUUUAAAGCCAAUUUGCUAUGCUUCGUUUAU------------------------GCUGG .....(((((((((..((((((.....((....))....))))))..)))))))))...((.........------------------------))... ( -18.60) >DroEre_CAF1 58095 98 - 1 UCCUCACAA-AUGGUCUCGAAUUCUUUAAGAAAUUUGUUGUUUAAAGCCAAUUUGCUGUGGUUGGUUUCGCCUGGCUUUUCCUAUUUAUUUUUUGCUGG .....((((-((..(((..(.....)..))).))))))......(((((((((......)))))))))...(..((..................))..) ( -15.17) >DroYak_CAF1 63778 91 - 1 UCCUCACAAAAUGGUCUCCAAUUCUUUAAGAAAUUUGUUGCUUAAAGCCAAUUUGCUUUGGUUCAAUUCGCCUGGCUUUUCCUAUUUAUUU-------- ........((((((.........(((((((..........))))))).......(((..(((.......))).))).....))))))....-------- ( -11.00) >consensus UCCUCGCAAAUUGGUCUUGAAUUCUUUAAGAAAUUUGUUGUUUAAAGCCAAUUUGCUAUGCUUCGUUUCGCCUGG_UUUU__U__U________GCUGG .....((((((((((.((((((.....((....))....)))))).))))))))))........................................... (-10.22 = -12.22 + 2.00)

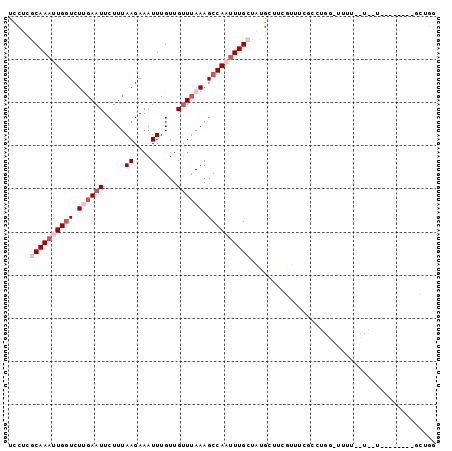
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:45 2006