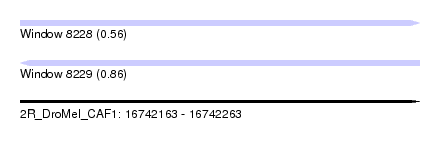
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,742,163 – 16,742,263 |
| Length | 100 |
| Max. P | 0.856585 |

| Location | 16,742,163 – 16,742,263 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
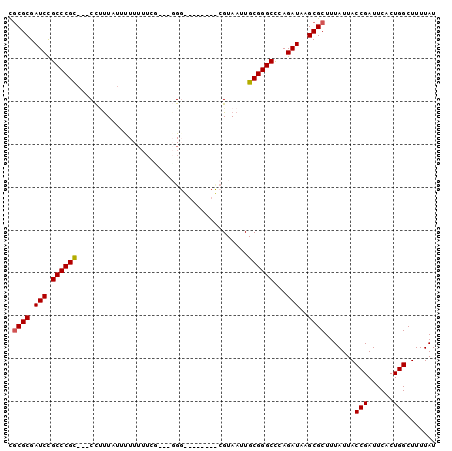
>2R_DroMel_CAF1 16742163 100 + 20766785 CGCGCGAUCCGCCCGUGCUCCCCUUUUCUUUUUCGAAUGGGGGUUCCCUCGUAAUUGCGGGCCCAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU .((((.(((.(((((((..(((((.(((......))).)))))..)..........))))))...)))..)))).......(((......)))....... ( -28.50) >DroSec_CAF1 49957 86 + 1 CGCGCGAUCCGCCCGCGCCCCUUUAUUUUUUUUU-----G---------GGUAAUUGCGGGCCCAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU .((((.(((.(((((((.(((.............-----)---------))....)))))))...)))..)))).......(((......)))....... ( -24.72) >DroSim_CAF1 52545 91 + 1 CGCGCGAUCCGCCCGCGCCCCUUUAUUUUUUUUUUUUUGG---------CGUAAUUGCGGGCCCAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU .((((.(((.((((((((.((.................))---------.))....))))))...)))..)))).......(((......)))....... ( -21.63) >DroEre_CAF1 51491 92 + 1 CGCGCGAUCCGCCCGC---CCUUU--UUUUUUUGG---GGGGGAGGGCUCGUAAUUGCGGGCCCAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU .((((.(((..(((.(---((...--.......))---).))).((((((((....)))))))).)))..)))).......(((......)))....... ( -35.80) >DroYak_CAF1 57062 91 + 1 CGCGCGAUCCGCCCGC---CCAUUAUUUUUUUUGC---GGG---GCCCCCGUAAUUGCGGGCCCAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU .((((.(((.((((((---............((((---(((---...)))))))..))))))...)))..)))).......(((......)))....... ( -27.34) >DroAna_CAF1 68367 86 + 1 UUCGCGAUCUGCCCGC---CUUUU--UUUU---CGUUUCGGG------CCGUAAUUGCGGGCUGAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU ...(((.......)))---.....--....---.(((((((.------((((....)))).)))))))....(((................)))...... ( -18.19) >consensus CGCGCGAUCCGCCCGC___CCUUUAUUUUUUUUCG___GGG________CGUAAUUGCGGGCCCAGAUAAGCGCUUUAUUACCGAUUCACUGGCUUUUAU .((((.(((.((((((........................................))))))...)))..)))).......(((......)))....... (-17.02 = -17.05 + 0.03)

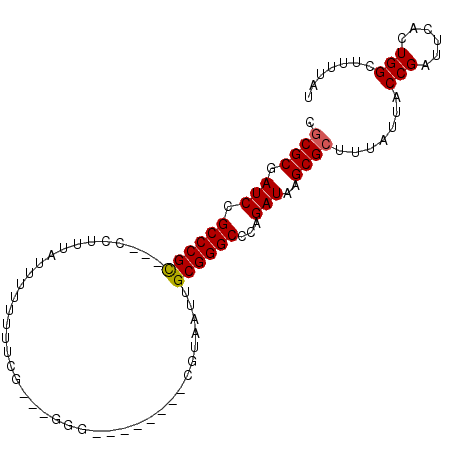
| Location | 16,742,163 – 16,742,263 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
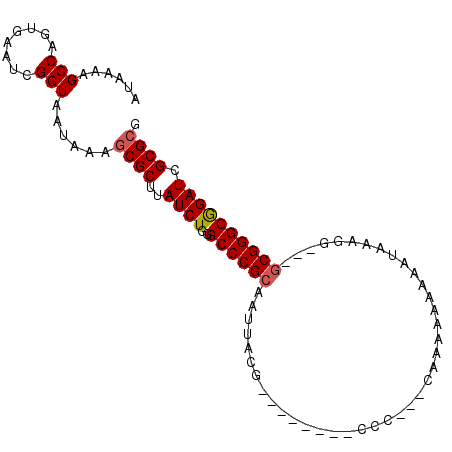
>2R_DroMel_CAF1 16742163 100 - 20766785 AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUGGGCCCGCAAUUACGAGGGAACCCCCAUUCGAAAAAGAAAAGGGGAGCACGGGCGGAUCGCGCG ......(((........)))......((((..((((..(((((.........(....)((((.(((......)))..))))....))))))))).)))). ( -28.80) >DroSec_CAF1 49957 86 - 1 AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUGGGCCCGCAAUUACC---------C-----AAAAAAAAAUAAAGGGGCGCGGGCGGAUCGCGCG ......(((........)))......((((..((((..((((((.....((---------(-----.............)))..)))))))))).)))). ( -27.12) >DroSim_CAF1 52545 91 - 1 AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUGGGCCCGCAAUUACG---------CCAAAAAAAAAAAAAUAAAGGGGCGCGGGCGGAUCGCGCG ......(((........)))......((((..((((..((((((......(---------((...................))))))))))))).)))). ( -24.91) >DroEre_CAF1 51491 92 - 1 AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUGGGCCCGCAAUUACGAGCCCUCCCCC---CCAAAAAAA--AAAGG---GCGGGCGGAUCGCGCG ......(((........)))......((((..((((((((.((......)).))))..((((---((.......--...))---).))).)))).)))). ( -29.20) >DroYak_CAF1 57062 91 - 1 AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUGGGCCCGCAAUUACGGGGGC---CCC---GCAAAAAAAAUAAUGG---GCGGGCGGAUCGCGCG ......(((........)))......((((..((((((((((.(......).))))---)((---((..(........)..---)))).))))).)))). ( -31.50) >DroAna_CAF1 68367 86 - 1 AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUCAGCCCGCAAUUACGG------CCCGAAACG---AAAA--AAAAG---GCGGGCAGAUCGCGAA ..........((((.((.(((((...(.(((......))).)...))))).(------((((...(.---....--....)---.))))).))))))... ( -19.90) >consensus AUAAAAGCCAGUGAAUCGGUAAUAAAGCGCUUAUCUGGGCCCGCAAUUACG________CCC___CAAAAAAAAAUAAAGG___GCGGGCGGAUCGCGCG ......(((........)))......((((..((((..((((((........................................)))))))))).)))). (-19.62 = -19.82 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:42 2006