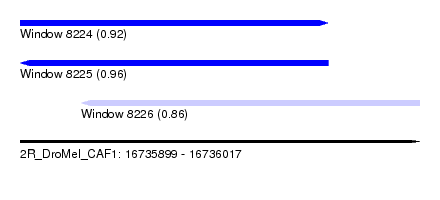
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,735,899 – 16,736,017 |
| Length | 118 |
| Max. P | 0.962161 |

| Location | 16,735,899 – 16,735,990 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
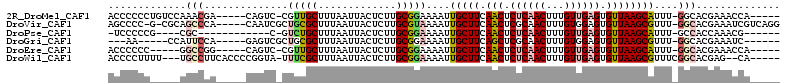
>2R_DroMel_CAF1 16735899 91 + 20766785 -UAUAUAUUUUUUUUUUUU---------------UGGUUUCGUGCCAAAUGCUUAACACUCAACAAAGUUGAGAGUUGAAGCAAUUUUCCGCAAGAGUAAUUAAAGC -....(((((((.....((---------------((((.....))))))((((((((.((((((...)))))).)))).)))).........)))))))........ ( -20.60) >DroVir_CAF1 65216 97 + 1 UUUG-----C-----UUUGGGGUCAGCCGCCUGACGAUUUCGUGCCAAACGCUUAACACUCCACAAAGUUGCGAGUUGAAGCAAUUUUACGCAAGAGUAAUUAAAGC ((((-----(-----(((((.(((((....)))))(....)...))))).((((...((((((......)).))))..))))........)))))............ ( -21.40) >DroPse_CAF1 45150 72 + 1 -----------------------------------CGUUUGGUGGCAAAUGCUUAACACUCAACAAAGUUGAGAGUUGAAGCAAUUUUCCGCAAGAGUAAUUAAAGC -----------------------------------......((((.(((((((((((.((((((...)))))).)))).))).)))).))))............... ( -19.70) >DroSec_CAF1 43868 85 + 1 -UGU-----UUUU-UUUUU---------------UGGUUUCGUGCCAAAUGCUUAACACUCAACAAAGUUGAGAGUUGAAGCAAUUUUCCGCAAGAGUAAUUAAAGC -...-----....-...((---------------((((.....))))))((((((((.((((((...)))))).)))).))))......(....)............ ( -19.60) >DroYak_CAF1 50693 81 + 1 -UAU-----U-----UUUU---------------UGGUUUCGUGCCAAAUGCUUAACACUCAACAAAGUUGAGAGUUGAAGCAAUUUUCCGCAAGAGUAAUUAAAGC -...-----.-----..((---------------((((.....))))))((((((((.((((((...)))))).)))).))))......(....)............ ( -19.60) >DroPer_CAF1 44314 72 + 1 -----------------------------------CGUUUGGUGGCAAAUGCUUAACACUCAACAAAGUUGAGAGUUGAAGCAAUUUUCCGCAAGAAUAAUUAAAGC -----------------------------------......((((.(((((((((((.((((((...)))))).)))).))).)))).))))............... ( -19.70) >consensus _U_U_____U_____UUUU_______________UGGUUUCGUGCCAAAUGCUUAACACUCAACAAAGUUGAGAGUUGAAGCAAUUUUCCGCAAGAGUAAUUAAAGC .........................................(((..(((((((((((.((((((...)))))).)))).))).))))..)))............... (-13.48 = -13.98 + 0.50)


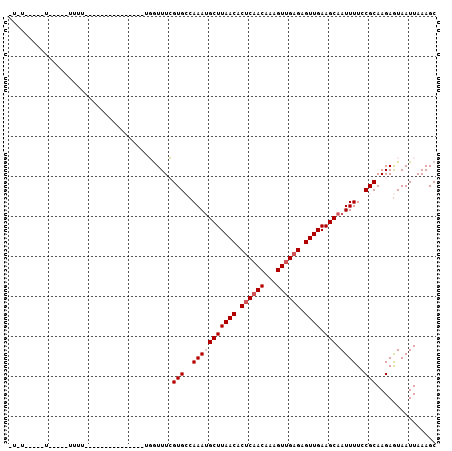
| Location | 16,735,899 – 16,735,990 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -19.72 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.65 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16735899 91 - 20766785 GCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUUGGCACGAAACCA---------------AAAAAAAAAAAAUAUAUA- ...........((.(((.(((...(((((.(((.((((((...)))))).))))))))))).))).)).....---------------..................- ( -19.60) >DroVir_CAF1 65216 97 - 1 GCUUUAAUUACUCUUGCGUAAAAUUGCUUCAACUCGCAACUUUGUGGAGUGUUAAGCGUUUGGCACGAAAUCGUCAGGCGGCUGACCCCAAA-----G-----CAAA (((((....(((((....((((.((((........)))).)))).)))))((((..((((((((........))))))))..))))...)))-----)-----)... ( -25.10) >DroPse_CAF1 45150 72 - 1 GCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUUGCCACCAAACG----------------------------------- ((.............))((.....(((((.(((.((((((...)))))).))))))))....))........----------------------------------- ( -17.22) >DroSec_CAF1 43868 85 - 1 GCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUUGGCACGAAACCA---------------AAAAA-AAAA-----ACA- ...........((.(((.(((...(((((.(((.((((((...)))))).))))))))))).))).)).....---------------.....-....-----...- ( -19.60) >DroYak_CAF1 50693 81 - 1 GCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUUGGCACGAAACCA---------------AAAA-----A-----AUA- ...........((.(((.(((...(((((.(((.((((((...)))))).))))))))))).))).)).....---------------....-----.-----...- ( -19.60) >DroPer_CAF1 44314 72 - 1 GCUUUAAUUAUUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUUGCCACCAAACG----------------------------------- ((.............))((.....(((((.(((.((((((...)))))).))))))))....))........----------------------------------- ( -17.22) >consensus GCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUUGGCACGAAACCA_______________AAAA_____A_____A_A_ ...........((.(((.(((...(((((.(((.((((((...)))))).))))))))))).))).))....................................... (-13.62 = -14.65 + 1.03)


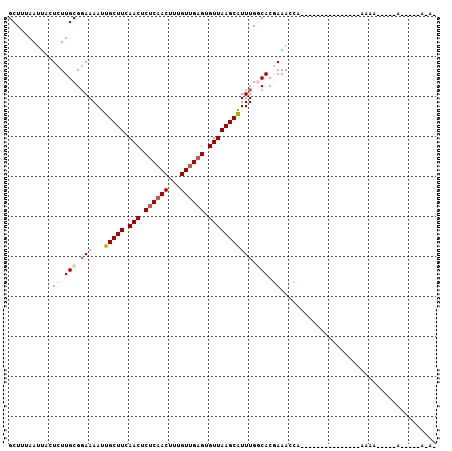
| Location | 16,735,917 – 16,736,017 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.10 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16735917 100 - 20766785 ACCCCCCUGUCCAAACGA-----CAGUC-CGUUGCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUU-GGCACGAAACCA----- ......(((((.....))-----)))..-...............((.(((.(((...(((((.(((.((((((...)))))).)))))))))))-.))).)).....----- ( -24.60) >DroVir_CAF1 65235 104 - 1 AGCCCC-G-CGCAGCCCA-----CAAUCGCUGCGCUUUAAUUACUCUUGCGUAAAAUUGCUUCAACUCGCAACUUUGUGGAGUGUUAAGCGUUU-GGCACGAAAUCGUCAGG .(((..-(-((((((...-----.....)))))((((....((((((....((((.((((........)))).)))).))))))..))))))..-)))(((....))).... ( -30.20) >DroPse_CAF1 45150 87 - 1 -UCCCCCG----CGC------------C-GUCUGCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUU-GCCACCAAACG------ -......(----(..------------.-.(((((.............)))))....(((((.(((.((((((...)))))).))))))))...-)).........------ ( -20.32) >DroGri_CAF1 62108 92 - 1 ---AA-----CCAUUCCA-----GAGUCGCUGCGCUUUAAUUACUCUUGCGGAAAAUUGCUUCAGCUCGCAACUUUGUGGAGUGUUAAGCGUUU-GGCACGAAAUC------ ---..-----........-----..((((..((((((....((((((.(((((...((((........)))).)))))))))))..)))))).)-)))........------ ( -22.30) >DroEre_CAF1 45281 95 - 1 ACCCCCC-----GGCCGG-----CAGUC-CGUUGCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUU-GGCACGAAACCA----- ......(-----((((((-----...((-(((................)))))....(((((.(((.((((((...)))))).)))))))).))-))).))......----- ( -26.09) >DroWil_CAF1 59127 101 - 1 ACCCCUUUU---UGCCUUCACCCCGGUA-UUUCGCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCGUUUCGGCACGAG--CA----- .........---((((........))))-..............(((.(((.((((...((((.(((.((((((...)))))).))))))).)))).))).)))--..----- ( -27.30) >consensus ACCCCCC_____AGCCCA_____CAGUC_CUUCGCUUUAAUUACUCUUGCGGAAAAUUGCUUCAACUCUCAACUUUGUUGAGUGUUAAGCAUUU_GGCACGAAACCA_____ .............(((..............(((((.............)))))....(((((.(((.((((((...)))))).))))))))....))).............. (-13.55 = -14.52 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:39 2006