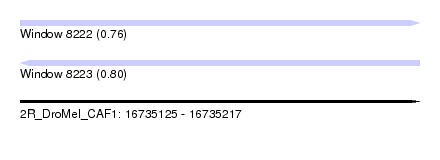
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,735,125 – 16,735,217 |
| Length | 92 |
| Max. P | 0.804852 |

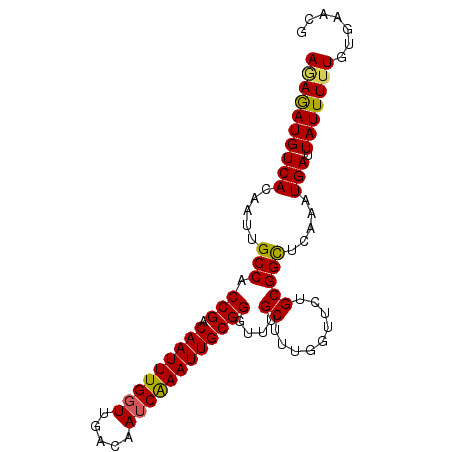
| Location | 16,735,125 – 16,735,217 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16735125 92 + 20766785 CGUUCACAAAAAUAAUCAUUUCAGCCGCAGAACCAAAGCGAACCCGCAAUUUGAUUGUCAACCAAAUUGUCGGUGGCAAUUGUGACAUCUCU .((.(((((((((....))))..(((((.........))..(((.((((((((.........)))))))).))))))..)))))))...... ( -20.30) >DroSec_CAF1 43105 92 + 1 CGUCCACAAAAAUAAUCAUUUGAGCCGCACAACCAAAGCAAACCCGCAAUUUGAUUGUCAACCAAAUUGUCGGUGGCAAUUGUGACAUCUCU .(((.((((......((....))(((((.........))..(((.((((((((.........)))))))).))))))..)))))))...... ( -22.00) >DroSim_CAF1 45410 92 + 1 CGUUCACAAAAAUAAUCAUUUGAGCCGCAGAACCAAAGCAAACCCGCAAUUUGAUUGUCAACCAAAUUGUCGGUGGCAAUUGUGACAUCUUU .((.(((((......((....))(((((.........))..(((.((((((((.........)))))))).))))))..)))))))...... ( -22.00) >DroEre_CAF1 44506 89 + 1 ---UCACAAAAAUAAUCAUUUCAACCGCCGAACCAAAGCAAACCCGCAAUUUGAUUGUCAAGCAAAUUGUCGGCGGCAAUUGUGACAUUUGU ---..(((((.....((((.....(((((((......((......))((((((.((....)))))))).))))))).....))))..))))) ( -20.20) >DroYak_CAF1 49885 92 + 1 CGUUUACAAAAAUAAUCAUUUAAACCGCAGAACCAAAGCAAACCCGCAAUUCGAUUGUCAACCAAAUUGUCGGUGGCAAUUGUGACAUUUCU .........................(((.........((......)).....((((((((.((........)))))))))))))........ ( -13.40) >consensus CGUUCACAAAAAUAAUCAUUUCAGCCGCAGAACCAAAGCAAACCCGCAAUUUGAUUGUCAACCAAAUUGUCGGUGGCAAUUGUGACAUCUCU ...((((((((((....))))..(((((.........))..(((.((((((((.........)))))))).))))))..))))))....... (-14.88 = -15.72 + 0.84)

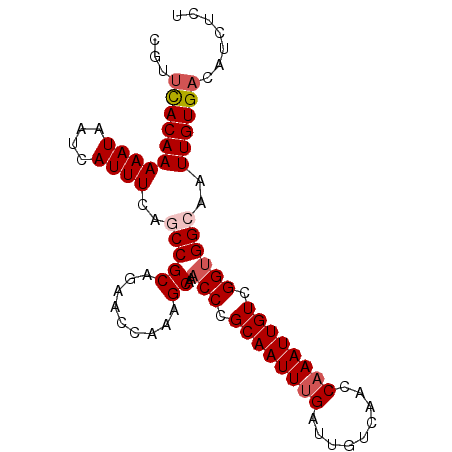
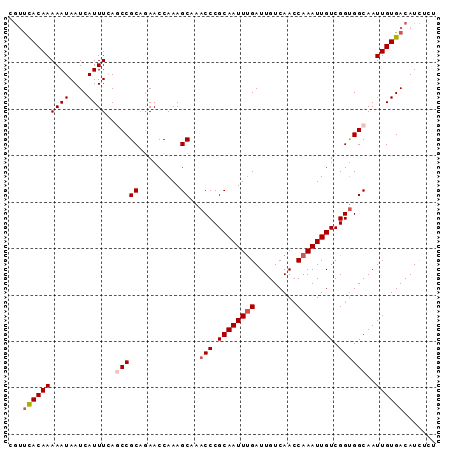
| Location | 16,735,125 – 16,735,217 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -21.80 |
| Energy contribution | -21.44 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16735125 92 - 20766785 AGAGAUGUCACAAUUGCCACCGACAAUUUGGUUGACAAUCAAAUUGCGGGUUCGCUUUGGUUCUGCGGCUGAAAUGAUUAUUUUUGUGAACG ((((((((((.....(((.(((.(((((((((.....))))))))))))....((.........))))).....))).)))))))....... ( -23.50) >DroSec_CAF1 43105 92 - 1 AGAGAUGUCACAAUUGCCACCGACAAUUUGGUUGACAAUCAAAUUGCGGGUUUGCUUUGGUUGUGCGGCUCAAAUGAUUAUUUUUGUGGACG .(((.((.(((((((((..(((.(((((((((.....))))))))))))....))...))))))))).)))..................... ( -24.20) >DroSim_CAF1 45410 92 - 1 AAAGAUGUCACAAUUGCCACCGACAAUUUGGUUGACAAUCAAAUUGCGGGUUUGCUUUGGUUCUGCGGCUCAAAUGAUUAUUUUUGUGAACG .......((((((..(((.(((.(((((((((.....))))))))))))....((.........)))))((....))......))))))... ( -23.40) >DroEre_CAF1 44506 89 - 1 ACAAAUGUCACAAUUGCCGCCGACAAUUUGCUUGACAAUCAAAUUGCGGGUUUGCUUUGGUUCGGCGGUUGAAAUGAUUAUUUUUGUGA--- (((((.((((.....(((((((((((...(((((.((((...))))))))).....)))..)))))))).....))))....)))))..--- ( -25.80) >DroYak_CAF1 49885 92 - 1 AGAAAUGUCACAAUUGCCACCGACAAUUUGGUUGACAAUCGAAUUGCGGGUUUGCUUUGGUUCUGCGGUUUAAAUGAUUAUUUUUGUAAACG ((((((((((.....(((.(((.(((((((((.....))))))))))))....((.........))))).....))).)))))))....... ( -21.20) >consensus AGAGAUGUCACAAUUGCCACCGACAAUUUGGUUGACAAUCAAAUUGCGGGUUUGCUUUGGUUCUGCGGCUCAAAUGAUUAUUUUUGUGAACG ((((((((((.....(((.(((.(((((((((.....))))))))))))....((.........))))).....))).)))))))....... (-21.80 = -21.44 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:37 2006