
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,728,857 – 16,728,968 |
| Length | 111 |
| Max. P | 0.959142 |

| Location | 16,728,857 – 16,728,968 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -52.68 |
| Consensus MFE | -32.33 |
| Energy contribution | -31.42 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
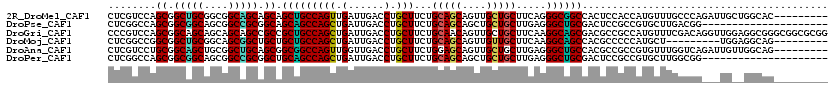
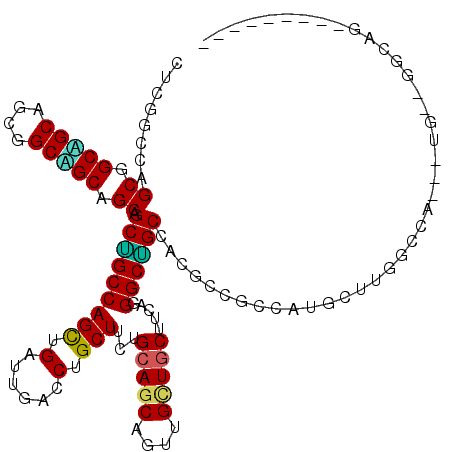
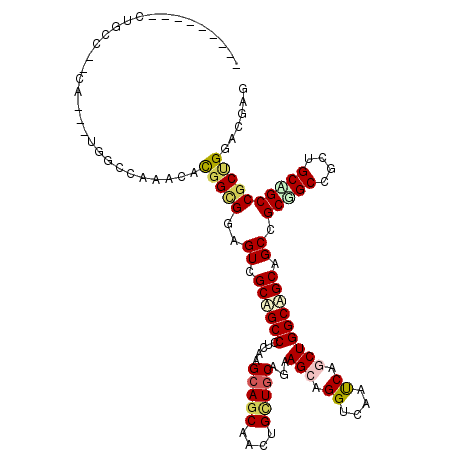
>2R_DroMel_CAF1 16728857 111 + 20766785 CUCGUCCAGCGGCUGCGGCGGCAGCAGCAGCUGCCAGUUGAUUGACCUGCUUCUGCAGCAGUUGCUGCUUCAGGGCGGCCACUCCACCAUGUUUGCCCAGAUUGCUGGCAC--------- ...(.((((((((((.(((((((((....)))))).((.((.((.((.(((.(((.(((((...))))).)))))))).)).)).)).......)))))).))))))))..--------- ( -49.50) >DroPse_CAF1 39624 99 + 1 CUCGGCCAGCGGCGGCAGCGGCCGCGGCAGCAGCCAGCUGAUUGACCUGCUUCUGCAGCAGCUGCUGCUUGAGGGCUGCGACUCCGCCGUGCUUGACGG--------------------- .(((((..(((((((..((((((.((((((((((..((((...((......))..)))).)))))))).))..))))))....))))))))).)))...--------------------- ( -51.00) >DroGri_CAF1 51962 120 + 1 CCCGUCCAGCGGCAGCAGCAGCAGCCGCCGCUGCCAGCUGAUUGACCUGCUUCUGCAACAGUUGCUGCUUCAAGGCAGCGACGCCGCCAUGUUUCGACAGGUUGGAGGCGGGCGGCGCGG ...(((((((((((((.((.......)).)))))).))))...)))......((((....(((((((((....)))))))))((((((.((((((.(.....).)))))))))))))))) ( -60.00) >DroMoj_CAF1 51409 102 + 1 CUCGGCCGGCGGCUGCGGCAGCGGCUGCUGCUGCCAGCUGAUUGACCUGCUUCUGCAGCAGUUGUUGCUUCAAGGCAGCCACGCCCCCAUGCU---------UGGAGGCAG--------- ....(((((((((((((((((((((((((((....(((.(......).)))...))))))))))))))).....))))))..))).(((....---------))).)))..--------- ( -51.60) >DroAna_CAF1 54280 111 + 1 CUCGUCCUGCGGCAGCUGCGGCUGCAGCGGCGGCCAGUUGGUUGACCUGCUUCUGGAGCAGUUGCUGCUUGAGGGCUGCCACGCCGCCGUGUUUGGUCAGAUUGUUGGCAG--------- ...((((((((((((((((((((((....))))))(((.((....)).)))......)))))))))))...)))))(((((((..((((....)))).....)).))))).--------- ( -50.70) >DroPer_CAF1 38780 99 + 1 CUCGGCCAGCGGCGGCAGCGGCCGCGGCUGCAGCCAGCUGAUUGACCUGCUUCUGCAGCAGCUGCUGCUUGAGGGCUGCGACUCCGCCGUGCUUGGCGG--------------------- (.((((((((.(((((....))))).)))(((((.(((((......((((....)))))))))))))).....))))).)...((((((....))))))--------------------- ( -53.30) >consensus CUCGGCCAGCGGCAGCAGCGGCAGCAGCAGCUGCCAGCUGAUUGACCUGCUUCUGCAGCAGUUGCUGCUUCAGGGCUGCCACGCCGCCAUGCUUGGCCA___UG__GGCAG_________ ........((.(((((....))))).)).(((((((((.(......).)))...(((((....))))).....))))))......................................... (-32.33 = -31.42 + -0.91)
| Location | 16,728,857 – 16,728,968 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -49.07 |
| Consensus MFE | -30.92 |
| Energy contribution | -31.37 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
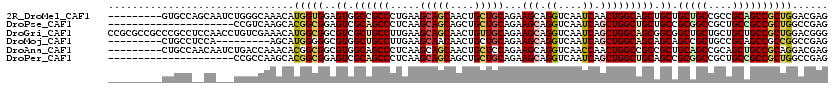
>2R_DroMel_CAF1 16728857 111 - 20766785 ---------GUGCCAGCAAUCUGGGCAAACAUGGUGGAGUGGCCGCCCUGAAGCAGCAACUGCUGCAGAAGCAGGUCAAUCAACUGGCAGCUGCUGCUGCCGCCGCAGCCGCUGGACGAG ---------((.(((((...((((((......(((.((.(((((((.(((.(((((...))))).)))..)).))))).)).)))((((((....))))))))).)))..)))))))... ( -51.80) >DroPse_CAF1 39624 99 - 1 ---------------------CCGUCAAGCACGGCGGAGUCGCAGCCCUCAAGCAGCAGCUGCUGCAGAAGCAGGUCAAUCAGCUGGCUGCUGCCGCGGCCGCUGCCGCCGCUGGCCGAG ---------------------..((((.((.((((((.(((((.((.....(((((((((((((((....))))......))))).)))))))).)))))..))))))..)))))).... ( -46.70) >DroGri_CAF1 51962 120 - 1 CCGCGCCGCCCGCCUCCAACCUGUCGAAACAUGGCGGCGUCGCUGCCUUGAAGCAGCAACUGUUGCAGAAGCAGGUCAAUCAGCUGGCAGCGGCGGCUGCUGCUGCUGCCGCUGGACGGG ((((((((((((.(........).))......)))))))).(((((......)))))..(((((.....)))))(((...((((.(((((((((((...)))))))))))))))))))). ( -57.60) >DroMoj_CAF1 51409 102 - 1 ---------CUGCCUCCA---------AGCAUGGGGGCGUGGCUGCCUUGAAGCAACAACUGCUGCAGAAGCAGGUCAAUCAGCUGGCAGCAGCAGCCGCUGCCGCAGCCGCCGGCCGAG ---------..(((((((---------....)))))))((((((((.....(((.....(((((.....)))))........)))((((((.......))))))))))))))........ ( -47.82) >DroAna_CAF1 54280 111 - 1 ---------CUGCCAACAAUCUGACCAAACACGGCGGCGUGGCAGCCCUCAAGCAGCAACUGCUCCAGAAGCAGGUCAACCAACUGGCCGCCGCUGCAGCCGCAGCUGCCGCAGGACGAG ---------((((((......)).........(((((((((((.(....)..(((((...((((.....))))(((((......)))))...))))).))))).))))))))))...... ( -42.20) >DroPer_CAF1 38780 99 - 1 ---------------------CCGCCAAGCACGGCGGAGUCGCAGCCCUCAAGCAGCAGCUGCUGCAGAAGCAGGUCAAUCAGCUGGCUGCAGCCGCGGCCGCUGCCGCCGCUGGCCGAG ---------------------(((((......)))))..(((..(((.....((((((((((((((....))))......))))).)))))(((.(((((....))))).))))))))). ( -48.30) >consensus _________CUGCC__CA___UGGCCAAACACGGCGGAGUCGCAGCCCUCAAGCAGCAACUGCUGCAGAAGCAGGUCAAUCAGCUGGCAGCAGCCGCGGCCGCUGCAGCCGCUGGACGAG ...............................(((((..((.((((((.....(((((....)))))...(((.((....)).))))))))).)).(((((....))))))))))...... (-30.92 = -31.37 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:34 2006