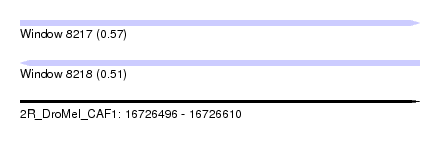
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,726,496 – 16,726,610 |
| Length | 114 |
| Max. P | 0.566069 |

| Location | 16,726,496 – 16,726,610 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -32.54 |
| Energy contribution | -32.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
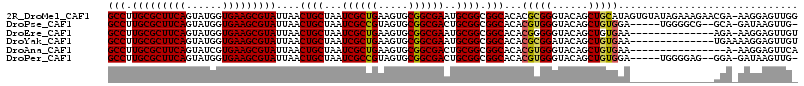
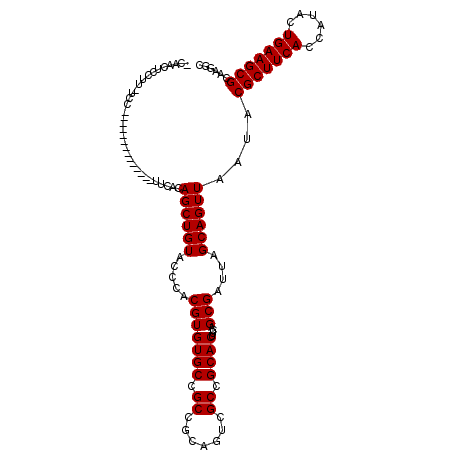
>2R_DroMel_CAF1 16726496 114 + 20766785 GCCUUGCGCUUCAGUAUGGUGAAGCGUAUUAACUGCUAAUCGCUGAAGUGCGGCGAAUGCGGCGGCACACGCGGGUACAGCUGCAUAGUGUAUAGAAAGAACGA-AAGGAGUUGG (((.(((((((((......)))))))))....((((...((((((.....))))))..)))).)))((((((((......))))...))))..........(..-..)....... ( -38.20) >DroPse_CAF1 37185 106 + 1 GCCUUGCGCUUCAGUAUGGUGAAGCGUAUUAACUGCUAAUCGCCGUAGUGCGGCGACUGCGGCGGCACACGUGGGUACAGCUGUGGA-----UGGGGCG--GCA-GAUAAGUUG- (((.(((((((((......)))))))))......(((.(((((((((((......))))))))..((((..((....))..))))))-----)..))))--)).-.........- ( -41.70) >DroEre_CAF1 35435 100 + 1 GCCUUGCGCUUCAGUAUGGUGAAGCGUAUUAACUGCUAAUCGCUGAAGUGCGGCGAAUGCGGCGGCACACGGGGGUACAGCUGUGAA--------------AGA-AAGGAGUUGU (((.(((((((((......)))))))))....((((...((((((.....))))))..)))).))).(((((........)))))..--------------...-.......... ( -35.30) >DroYak_CAF1 41037 101 + 1 GCCUUGCGCUUCAGUAUGGUGAAGCGUAUUAACUGCUAAUCGCUGAAGUGCGGCGAAUGCGGCGGCACACGCGGAUACAGCUGUGAA--------------UGAAAAGGAGUUGU (((.(((((((((......)))))))))....((((...((((((.....))))))..)))).)))...(((((......)))))..--------------.............. ( -35.00) >DroAna_CAF1 51883 98 + 1 GCCUUGCGCUUCAGUAUCGUGAAGCGUAUUAACUGCUAAUCGCUGAAGUGCGGCGACUGCGGCGGCACACGUGGGUACAGCUGUGAA----------------A-AAGGAGUUCA (((.(((((((((......)))))))))....((((...((((((.....))))))..)))).))).((((((....))..))))..----------------.-.......... ( -32.50) >DroPer_CAF1 36334 106 + 1 GCCUUGCGCUUCAGUAUGGUGAAGCGUAUUAACUGCUAAUCGCCGUAGUGCGGCGACUGCGGCGGCACACGUGGGUACAGCUGUGGA-----UGGGGAG--GGA-GAUAAGUUG- .((.(((((((((......)))))))))....((.((.(((((((((((......))))))))..((((..((....))..))))))-----).)).))--)).-.........- ( -37.60) >consensus GCCUUGCGCUUCAGUAUGGUGAAGCGUAUUAACUGCUAAUCGCUGAAGUGCGGCGAAUGCGGCGGCACACGUGGGUACAGCUGUGAA______________GGA_AAGGAGUUG_ (((.(((((((((......)))))))))....((((...((((((.....))))))..)))).)))...(((((......))))).............................. (-32.54 = -32.35 + -0.19)
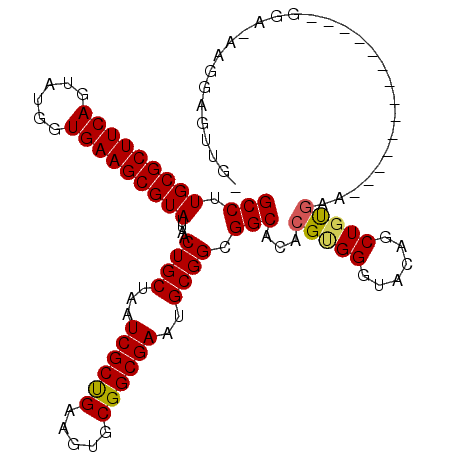
| Location | 16,726,496 – 16,726,610 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.57 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16726496 114 - 20766785 CCAACUCCUU-UCGUUCUUUCUAUACACUAUGCAGCUGUACCCGCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGC ..........-................((.(((((((((....(((.((((....)))).)))(((......)))....)))))).....((((((......))))))))).)). ( -29.40) >DroPse_CAF1 37185 106 - 1 -CAACUUAUC-UGC--CGCCCCA-----UCCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUACGGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGC -.........-.((--(......-----......((.((((......)))).)).((((((((((.....)))))))).))........(((((((......)))))))...))) ( -32.80) >DroEre_CAF1 35435 100 - 1 ACAACUCCUU-UCU--------------UUCACAGCUGUACCCCCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGC ......((((-...--------------.....((((((.....((.((((....)))).)).(((......)))....))))))....(((((((......))))))).)))). ( -24.80) >DroYak_CAF1 41037 101 - 1 ACAACUCCUUUUCA--------------UUCACAGCUGUAUCCGCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGC ......((((....--------------.....((((((....(((.((((....)))).)))(((......)))....))))))....(((((((......))))))).)))). ( -28.60) >DroAna_CAF1 51883 98 - 1 UGAACUCCUU-U----------------UUCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACGAUACUGAAGCGCAAGGC ......((((-.----------------......((.((((......)))).)).((((((((.........)))))).))........(((((((......))))))).)))). ( -24.30) >DroPer_CAF1 36334 106 - 1 -CAACUUAUC-UCC--CUCCCCA-----UCCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUACGGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGC -.........-...--.......-----..((((..((....))..))))..(((((((((((((.....)))))))).))........(((((((......)))))))...))) ( -30.60) >consensus _CAACUCCUU_UCC______________UUCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGC .................................((((((.....(((((((.((.......)).))))....)))....))))))....(((((((......)))))))...... (-22.57 = -22.57 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:32 2006