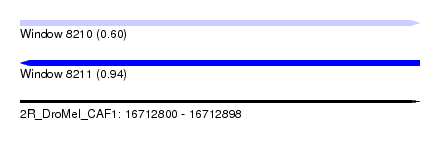
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,712,800 – 16,712,898 |
| Length | 98 |
| Max. P | 0.941960 |

| Location | 16,712,800 – 16,712,898 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16712800 98 + 20766785 GGCGGACAUUAAGAAGCAUUUGCCAUUUGUGUACAAUGUGACGGCUUAUGCCUCGUACUGUUAUCUGCCCGCCUUCAAACAGUAAGUUCGCUAUCU-CU------------ (((((((.....((.((((..(((..(((....)))......)))..)))).)).(((((((...............))))))).)))))))....-..------------ ( -26.46) >DroVir_CAF1 25236 103 + 1 CGCAGAUAUCAAGAAGCAUUUGCCCUUUGUCUAUAAUGUGACAGCCUAUGCUUCCUACUGCUACUUGCCUGCGUUCAAACAGUAAGUGGGCCAUGCACU-------GCA-A .((((.......(((((((..((....((((........))))))..)))))))....(((.....(((..(...(.....)...)..)))...)))))-------)).-. ( -30.60) >DroPse_CAF1 23211 108 + 1 GGCGGACAUCAAGAAGCAUUUGCCAUUCGUGUACAAUGUGACGGCCUAUGCCUCCUACUGCUAUCUGCCCGCCUUCAAACAGUAAGUCGACCAUCC-CAACCCCCUCCC-- (((((.......((.((((..(((..(((..(...)..))).)))..)))).)).....((.....))))))).......................-............-- ( -22.10) >DroMoj_CAF1 27106 102 + 1 CGCAGAUAUCAAGAAGCAUUUGCCAUUUGUCUACAACGUGACGGCCUAUGCUUCCUAUUGCUAUUUGCCCGCAUUCAAACAGUAAGUC---CGUGGAUGAU-----CUA-G .(((((((.((((((((((..(((....(((........))))))..)))))))...))).)))))))...((((((.((.....)).---..))))))..-----...-. ( -28.50) >DroAna_CAF1 26664 103 + 1 GGCGGACAUCAAGAAGCACUUGCCAUUUGUUUACAAUGUGACGGCCUAUGCCUCCUACUGCUAUCUGCCCGCCUUCAAACAGUAAGUUC-CUAUCA-CC------UCCCAG (((((......(((((((.....((((((....))).)))..(((....)))......)))).)))..)))))................-......-..------...... ( -18.80) >DroPer_CAF1 22479 108 + 1 GGCGGACAUCAAGAAGCAUUUGCCAUUCGUCUACAAUGUGACGGCCUAUGCCUCCUACUGCUAUCUGCCCGCCUUCAAACAGUAAGUUGACCAUCC-CAACCCCCUCCA-- (((((.......((.((((..(((....(((........))))))..)))).)).....((.....)))))))............((((.......-))))........-- ( -24.60) >consensus GGCGGACAUCAAGAAGCAUUUGCCAUUUGUCUACAAUGUGACGGCCUAUGCCUCCUACUGCUAUCUGCCCGCCUUCAAACAGUAAGUUG_CCAUCC_CA_______CCA__ (((((.......((.((((..(((....(((........))))))..)))).)).....((.....)))))))...................................... (-17.10 = -17.43 + 0.33)

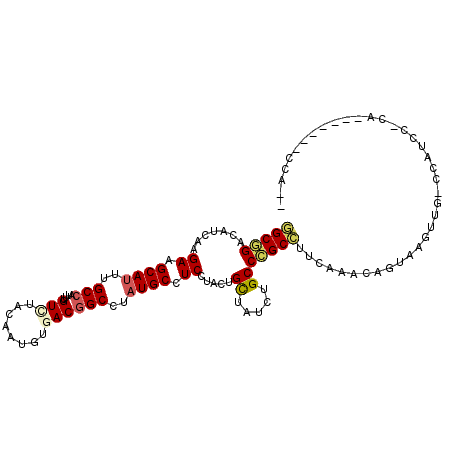
| Location | 16,712,800 – 16,712,898 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -28.52 |
| Energy contribution | -27.67 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16712800 98 - 20766785 ------------AG-AGAUAGCGAACUUACUGUUUGAAGGCGGGCAGAUAACAGUACGAGGCAUAAGCCGUCACAUUGUACACAAAUGGCAAAUGCUUCUUAAUGUCCGCC ------------..-......(((((.....)))))..((((((((........((.(((((((..(((((....(((....))))))))..))))))).)).)))))))) ( -31.70) >DroVir_CAF1 25236 103 - 1 U-UGC-------AGUGCAUGGCCCACUUACUGUUUGAACGCAGGCAAGUAGCAGUAGGAAGCAUAGGCUGUCACAUUAUAGACAAAGGGCAAAUGCUUCUUGAUAUCUGCG .-.((-------((......((..((((.((((......))))..)))).))..((((((((((..((((((........)))....)))..))))))))))....)))). ( -30.30) >DroPse_CAF1 23211 108 - 1 --GGGAGGGGGUUG-GGAUGGUCGACUUACUGUUUGAAGGCGGGCAGAUAGCAGUAGGAGGCAUAGGCCGUCACAUUGUACACGAAUGGCAAAUGCUUCUUGAUGUCCGCC --...((..(((((-(.....))))))..)).......((((((((........((((((((((..(((((....(((....))))))))..)))))))))).)))))))) ( -39.20) >DroMoj_CAF1 27106 102 - 1 C-UAG-----AUCAUCCACG---GACUUACUGUUUGAAUGCGGGCAAAUAGCAAUAGGAAGCAUAGGCCGUCACGUUGUAGACAAAUGGCAAAUGCUUCUUGAUAUCUGCG .-...-----........((---(((.....)))))...((((((.....))..((((((((((..(((((....(((....))))))))..))))))))))....)))). ( -29.30) >DroAna_CAF1 26664 103 - 1 CUGGGA------GG-UGAUAG-GAACUUACUGUUUGAAGGCGGGCAGAUAGCAGUAGGAGGCAUAGGCCGUCACAUUGUAAACAAAUGGCAAGUGCUUCUUGAUGUCCGCC ......------..-.(((((-.......)))))....((((((((........((((((((((..(((((....(((....))))))))..)))))))))).)))))))) ( -35.10) >DroPer_CAF1 22479 108 - 1 --UGGAGGGGGUUG-GGAUGGUCAACUUACUGUUUGAAGGCGGGCAGAUAGCAGUAGGAGGCAUAGGCCGUCACAUUGUAGACGAAUGGCAAAUGCUUCUUGAUGUCCGCC --...((..(((((-(.....))))))..)).......((((((((........((((((((((..(((((....(((....))))))))..)))))))))).)))))))) ( -39.50) >consensus __UGG_______AG_GGAUGG_CAACUUACUGUUUGAAGGCGGGCAGAUAGCAGUAGGAGGCAUAGGCCGUCACAUUGUAGACAAAUGGCAAAUGCUUCUUGAUGUCCGCC ......................................((((((((........((((((((((..(((((....(((....))))))))..)))))))))).)))))))) (-28.52 = -27.67 + -0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:25 2006