
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,693,733 – 16,693,847 |
| Length | 114 |
| Max. P | 0.858657 |

| Location | 16,693,733 – 16,693,847 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -26.88 |
| Energy contribution | -25.13 |
| Covariance contribution | -1.74 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16693733 114 + 20766785 UACUACAAUGCAAACCAGAGAGUUCCCGAUCAGUUGAUCUUGGGCGCACCCUGCAUUGUUAAAUGCGAUCAGGAAUGGUACCGUGCAGAGAUUCUGCGCGUUGAUGACUCUG---UG ...............(((((.(((((.((((..........(((....))).(((((....))))))))).))))).....(((((((.....))))))).......)))))---.. ( -37.60) >DroPse_CAF1 3396 114 + 1 UACUACAACAUCAACCAGAAGGUACCCGACAAACUCACCCUGGGUGCCCCCUGCAUCGUCAAAUGCGAUCAGGAGUGGUAUCGUGCAGAGAUCCUGCGUGUCGAUGACUCUG---UA ((((((..............((((((((............)))))))).((((.(((((.....))))))))).))))))...(((((((.((...((...))..)))))))---)) ( -36.50) >DroWil_CAF1 3818 117 + 1 UAUUACAACAUACAUCAGGUGGUGCCUGAUCAAUUGAUCUUGGGUGCUCCCUGCAUAGUGAAAUGCGAUCAGGAAUGGUAUCGGGCAGAGAUUCUACGUGUGGCUGAUUCCACAAUU ..................(((((((((((((..((((((..(((....))).((((......)))))))))).....).))))))))......)))).(((((......)))))... ( -32.20) >DroMoj_CAF1 2230 117 + 1 UACUAUAAUGUCAAUCAAGUGGUGCCCGAGCAGCUGGUGCUGGGUGCCCCGUGCGUCGUUAAAUGCGACCGCGAAUGGUAUCGUGCGGAGCUGCUGCGCAUAGAUGACACCACAAUC ........(((((((((...((..(((.((((.....)))))))..)).((((.(((((.....)))))))))..))))...((((((.....)))))).....)))))........ ( -43.10) >DroAna_CAF1 2064 114 + 1 UACUAUAAUGCAAACCAAAAAGUGCCUGACCAACUGGUCCUGGGUGCUCCGUGCAUCGUUAAAUGCGACCAGGAGUGGUACCGAGCGGAGAUCCUCCGCGUUGAUGACUCUG---UG ........((((........((..(((((((....))))..)))..))...))))((((.....))))...(((((.((.....((((((...))))))....)).))))).---.. ( -36.20) >DroPer_CAF1 2089 114 + 1 UACUACAACAUCAACCAGAAGGUACCCGACAAACUCACCCUGGGUGCCCCCUGCAUCGUCAAAUGCGAUCAGGAGUGGUAUCGUGCAGAGAUCCUGCGUGUCGAUGACUCUG---UA ((((((..............((((((((............)))))))).((((.(((((.....))))))))).))))))...(((((((.((...((...))..)))))))---)) ( -36.50) >consensus UACUACAACAUAAACCAGAAGGUGCCCGACCAACUGAUCCUGGGUGCCCCCUGCAUCGUUAAAUGCGAUCAGGAAUGGUAUCGUGCAGAGAUCCUGCGCGUCGAUGACUCUG___UA .......((((..((((...((((((((............)))))))).((((.(((((.....)))))))))..)))).....((((.....))))))))................ (-26.88 = -25.13 + -1.74)

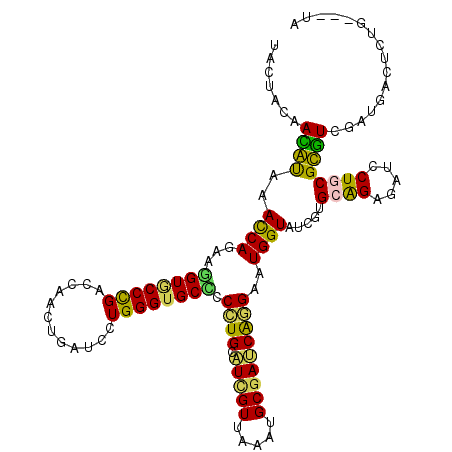
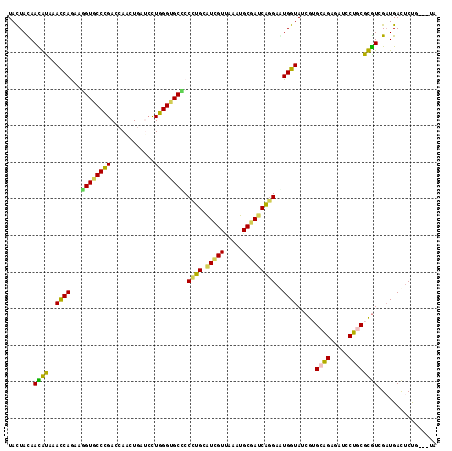
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:20 2006