| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,692,064 – 16,692,278 |
| Length | 214 |
| Max. P | 0.990779 |

| Location | 16,692,064 – 16,692,161 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -10.86 |
| Energy contribution | -11.00 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16692064 97 + 20766785 GA-ACAAUACCUU-UCUAAACAAUUUAUCCCUUUUAGUUAAUGUUCCCCAAGCAGCCCAAGCUGCUUCAGAACAUUCCCCUGCACUGCUUCCAGUACAU ..-..........-......................(..(((((((...(((((((....)))))))..)))))))..).((.((((....)))).)). ( -19.10) >DroPse_CAF1 1702 97 + 1 AGCAGAAUAUCUAACGUGUG--UUCUUUCUUUUCCAGUUAAUGUUCCUUAAGCAGCCGAAGCUUCUUCUGAACAUCCCCAUUCUCUGCUCACAGUACAU ((((((.....((((.....--..............))))((((((...(((.(((....))).)))..))))))........)))))).......... ( -15.71) >DroSec_CAF1 381 97 + 1 AA-ACAAUGCCUU-UCUAAACAAUAUAUCCCUUUUAGUUAAUGUUCCCCAAGCAGCCCAAGCUGCUGCAGAGUAUUCCCCUGCACUGCUUCCAGUACAU ..-..........-......................((...((.......((((((....))))))((((.(((......))).))))...))..)).. ( -16.40) >DroSim_CAF1 378 97 + 1 AA-ACAAUGCCUU-UCUAAACAAUUUAUCCCUUUUAGUUAAUGUUCCCCAAGCAGCCCAAGCUGCUGCAGAGCAUUCCCCUGCACUGCUUCCAGUACAU ..-..........-......................(..(((((((....((((((....))))))...)))))))..).((.((((....)))).)). ( -18.90) >DroYak_CAF1 380 98 + 1 AA-GCAAUUUCUUAUUUAUGAAAUCUAUCCUUUUUAGCUUAUGUUCCCGAAGCAACCCAAGCUGCUGCAGAACAUUCCCCUGCUCUGCUCCCAGUACAU .(-((.(((((........)))))...........(((((..(((........)))..))))))))(((((.((......)).)))))........... ( -16.20) >DroPer_CAF1 394 97 + 1 AGCAGAAUAUCUAACGUGUG--UUCUUUCUUUUCCAGUUAAUGUUCCUUAAGCAGCCGAAGCUUCUUCUGAACAUCCCCAUUCUCUGCUCACAGUACAU ((((((.....((((.....--..............))))((((((...(((.(((....))).)))..))))))........)))))).......... ( -15.71) >consensus AA_ACAAUACCUU_UCUAAACAAUCUAUCCCUUUUAGUUAAUGUUCCCCAAGCAGCCCAAGCUGCUGCAGAACAUUCCCCUGCACUGCUCCCAGUACAU ........................................((((((....((((((....))))))...)))))).........(((....)))..... (-10.86 = -11.00 + 0.14)


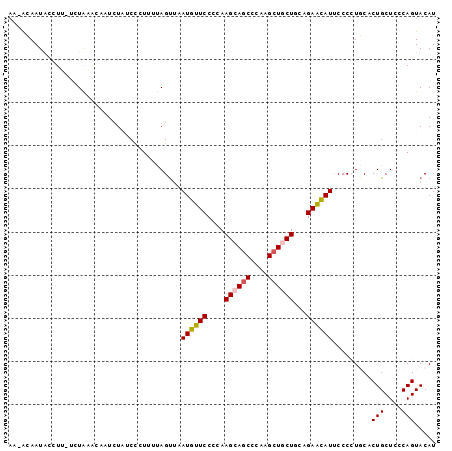
| Location | 16,692,064 – 16,692,161 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -18.09 |
| Energy contribution | -19.78 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16692064 97 - 20766785 AUGUACUGGAAGCAGUGCAGGGGAAUGUUCUGAAGCAGCUUGGGCUGCUUGGGGAACAUUAACUAAAAGGGAUAAAUUGUUUAGA-AAGGUAUUGU-UC .(((((((....)))))))((..((((((((.(((((((....))))))).).)))))))..))...((.((((..((.......-))..)))).)-). ( -32.60) >DroPse_CAF1 1702 97 - 1 AUGUACUGUGAGCAGAGAAUGGGGAUGUUCAGAAGAAGCUUCGGCUGCUUAAGGAACAUUAACUGGAAAAGAAAGAA--CACACGUUAGAUAUUCUGCU ..........(((((((...(..(((((((..(((.(((....))).)))...)))))))..)............((--(....))).....))))))) ( -26.10) >DroSec_CAF1 381 97 - 1 AUGUACUGGAAGCAGUGCAGGGGAAUACUCUGCAGCAGCUUGGGCUGCUUGGGGAACAUUAACUAAAAGGGAUAUAUUGUUUAGA-AAGGCAUUGU-UU .(((((((....)))))))((..(((..(((.(((((((....))))).)).)))..)))..)).....(((((...(((((...-.))))).)))-)) ( -27.90) >DroSim_CAF1 378 97 - 1 AUGUACUGGAAGCAGUGCAGGGGAAUGCUCUGCAGCAGCUUGGGCUGCUUGGGGAACAUUAACUAAAAGGGAUAAAUUGUUUAGA-AAGGCAUUGU-UU .(((((((....)))))))((..((((.(((.(((((((....))))).)).))).))))..)).....((((((..(((((...-.)))))))))-)) ( -31.30) >DroYak_CAF1 380 98 - 1 AUGUACUGGGAGCAGAGCAGGGGAAUGUUCUGCAGCAGCUUGGGUUGCUUCGGGAACAUAAGCUAAAAAGGAUAGAUUUCAUAAAUAAGAAAUUGC-UU ...........((((((((......))))))))(((((((((.(((.(....).))).))))))..........((((((........))))))))-). ( -30.50) >DroPer_CAF1 394 97 - 1 AUGUACUGUGAGCAGAGAAUGGGGAUGUUCAGAAGAAGCUUCGGCUGCUUAAGGAACAUUAACUGGAAAAGAAAGAA--CACACGUUAGAUAUUCUGCU ..........(((((((...(..(((((((..(((.(((....))).)))...)))))))..)............((--(....))).....))))))) ( -26.10) >consensus AUGUACUGGAAGCAGAGCAGGGGAAUGUUCUGAAGCAGCUUGGGCUGCUUGGGGAACAUUAACUAAAAAGGAUAGAUUGCAUAGA_AAGAUAUUGU_UU .(((.(((....))).)))((..((((((((..((((((....))))))...))))))))..))................................... (-18.09 = -19.78 + 1.70)

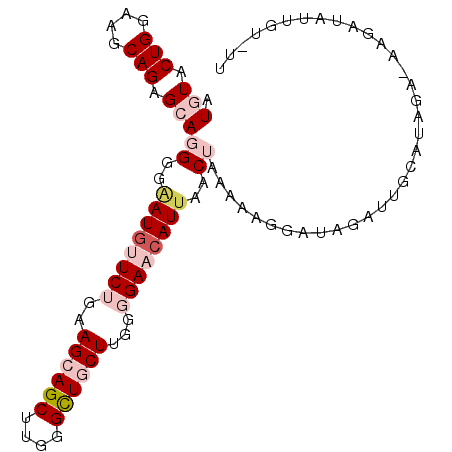
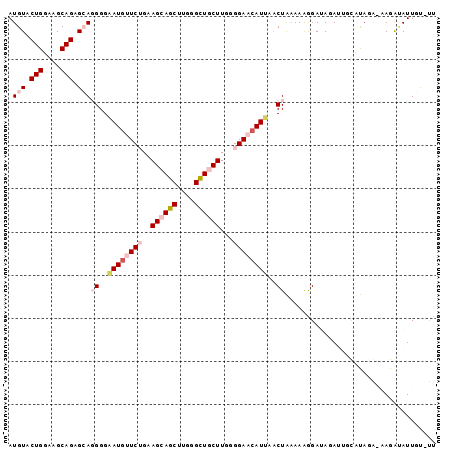
| Location | 16,692,093 – 16,692,198 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -35.33 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16692093 105 - 20766785 AUCG---GUCUGAUCCCACUCGGAGCAGAUUCCCAGCACAAUGUACUGGAAGCAGUGCAGGGGAAUGUUCUGAAGCAGCUUGGGCUGCUUGGGGAACAUUAACUAAAA-- ...(---((.((.((((...(((((((..(((((.......(((((((....))))))).))))))))))))(((((((....))))))).)))).))...)))....-- ( -42.50) >DroEre_CAF1 421 105 - 1 AUCG---GUCUGAUCCCACUCGGAGCAGAUGCCCAGCACAAUGUACUGGGAGCAAAGCAGGGGAAUGUUCUGCAGCAGCUUGGGUUGCUUGGGGAACAUAAGCUAAAA-- ....---(((((.(((.....))).))))).(((((.((...)).))))).....(((......(((((((.(((((((....))))).)).)))))))..)))....-- ( -34.00) >DroWil_CAF1 2113 108 - 1 AUCACUAGCCUUCUCCCAAUCAGAGCAAAUUCCCAGUACUAUAUAUUGUGAGCAGAGGAGGGGUAUAUUCUGUAGAAGCUUCGGUUGCUUCGAGAACAUCAACUGGAA-- ....(((((((((((....((((((....))).(((((.....))))))))...)))))))(((...((((...(((((.......))))).)))).)))..))))..-- ( -23.20) >DroYak_CAF1 410 105 - 1 AUCG---GUCUGAUCCCACUCGGAGCAGAUGCCCAGCACAAUGUACUGGGAGCAGAGCAGGGGAAUGUUCUGCAGCAGCUUGGGUUGCUUCGGGAACAUAAGCUAAAA-- ....---(((((.(((.....))).))))).(((((.((...)).))))).((((((((......))))))))...((((((.(((.(....).))).))))))....-- ( -41.00) >DroMoj_CAF1 467 107 - 1 UUCA---GUUUGGUCCCAUUCCGAGCAAAUGCCCAGCACAAUGUACUGCGAACAGAGCACAGGCACAUUUUGCAGCAGCUGCGGCUGUUUCGAGAAGAGCAGCUGAAAUA ((((---(((.(.((...(((.(((((...(((((((....((..(((((((..(.((....)).)..))))))))))))).)))))))).)))..)).))))))))... ( -31.00) >DroAna_CAF1 404 105 - 1 AUCG---GACUGAUCCCACUCGGCACAAAUGCCGAGGACUAUGUACUGCGUGCAGAGCAGUGGAAGGUUCUGUAGCAGCUUUGGCUGUUUGGGGAACAUUAACUGGGA-- ....---......((((((((((((....)))))))..((.(.((((((.......)))))).)))(((((.(((((((....)))).))).)))))......)))))-- ( -40.30) >consensus AUCG___GUCUGAUCCCACUCGGAGCAAAUGCCCAGCACAAUGUACUGGGAGCAGAGCAGGGGAAUGUUCUGCAGCAGCUUGGGCUGCUUCGGGAACAUAAACUAAAA__ .............((((...(((((((....(((.......(((.(((....))).))).)))..))))))).((((((....))))))..))))............... (-20.92 = -21.73 + 0.81)

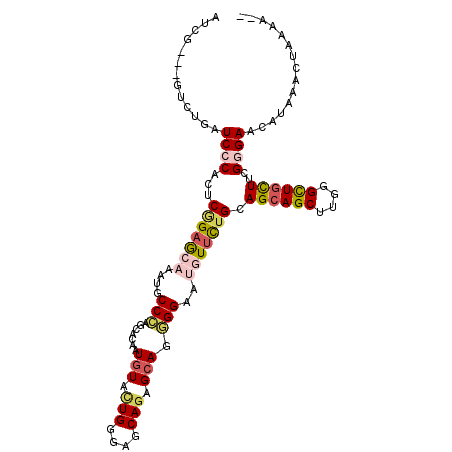
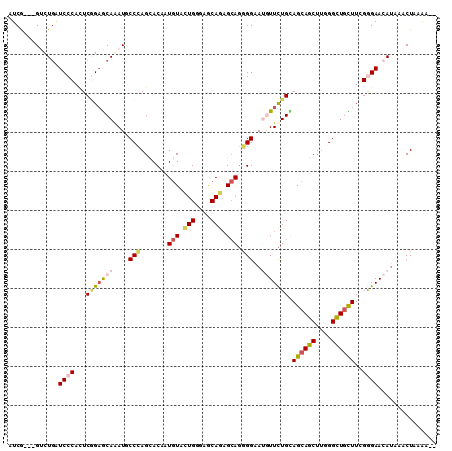
| Location | 16,692,161 – 16,692,278 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -18.16 |
| Energy contribution | -21.11 |
| Covariance contribution | 2.95 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16692161 117 - 20766785 AACUUUUGGUCACAGAUUUGUGUAGGUUCCACAGUGAUUUUAACGAUCUGGUUGACAACCAGCCGCCGAACCUCAGCCAGAUCGGUCUGAUCCCACUCGGAGCAGAUUCCCAGCACA ....((((....))))..(((((.((.................((((((((((((.................))))))))))))(((((.(((.....))).)))))..)).))))) ( -34.43) >DroVir_CAF1 490 117 - 1 AAUUUUUGGCCACAGAUUUGUGUGCUAUCCACCUUGAUGCUGACCACAUGAUUUACCAUCAGCUGCUUCACUGCAUCCAGCUCGGACUGAUCCCAUUCCGAGCAAAUGCCGAGCACA ......(((((((......))).))))...........(((((.....((......)))))))(((((....((((...(((((((.((....)).)))))))..)))).))))).. ( -32.30) >DroEre_CAF1 489 117 - 1 AACUUUUGGUCACAGAUAUGUGUGGGAUCAACGGUGAUUUGAACGGUCUGGUUGACAAUCAGAUCCUGAACCACAGCCAGAUCGGUCUGAUCCCACUCGGAGCAGAUGCCCAGCACA ....((((....))))...(.(((((((((.(((....)))..(((((((((((....((((...))))....)))))))))))...))))))))).)((.((....))))...... ( -41.30) >DroYak_CAF1 478 117 - 1 AAUUUUUGGUCACAAAUAUGUGUGGGAUCAACGUUGAUUUGAACGGUCUGGUUGACAAUCAGAUCUUGAACCACAGCCAGAUCGGUCUGAUCCCACUCGGAGCAGAUGCCCAGCACA ....((((....))))...(.((((((((((((((......)))((((((((((....((((...))))....)))))))))).)).))))))))).)((.((....))))...... ( -39.00) >DroMoj_CAF1 537 117 - 1 AACUUUUGACCACAGAUCUGUGUGGGAUCCACCUCAAUGCUGACCACAUGGUUAACCAUCAGCUGUUUCACAGUAUCCAGUUCAGUUUGGUCCCAUUCCGAGCAAAUGCCCAGCACA ...................(((((((......(((((((..(((((.((((....))))..((((.....)))).............))))).))))..)))......)))).))). ( -26.40) >DroAna_CAF1 472 117 - 1 AACUUCUGGCCACAGAUCUGCGUCGGAUCGAUGUCGAUGUGGACGGUCUGGUUGACGAUCAGCUUCUGGACCACUGCUAGAUCGGACUGAUCCCACUCGGCACAAAUGCCGAGGACU ....((((....)))).....(((((((((...(((((.(((..((((((((((.....)))))...)))))....))).)))))..))))))..(((((((....)))))))))). ( -46.60) >consensus AACUUUUGGCCACAGAUAUGUGUGGGAUCCACGUUGAUGUGAACGAUCUGGUUGACAAUCAGCUGCUGAACCACAGCCAGAUCGGUCUGAUCCCACUCGGAGCAAAUGCCCAGCACA ....((((....))))...(((((((.................(((((((((((...................)))))))))))(((((.(((.....))).))))).)))).))). (-18.16 = -21.11 + 2.95)

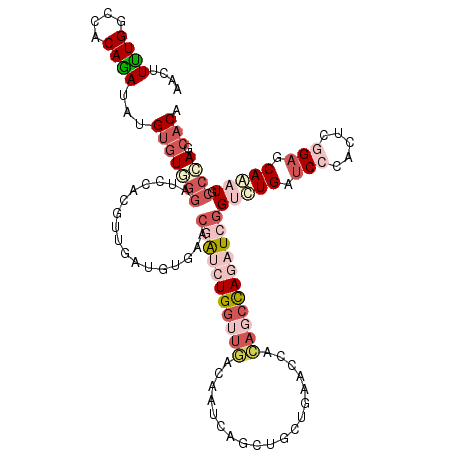
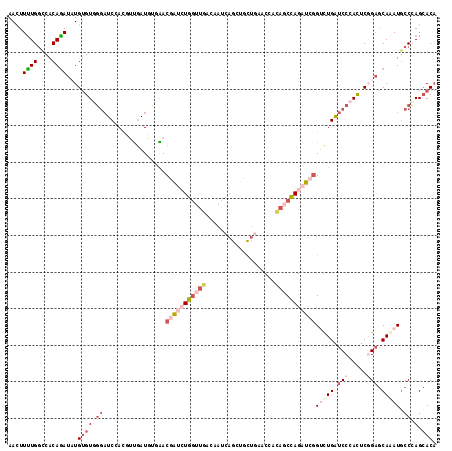
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:18 2006