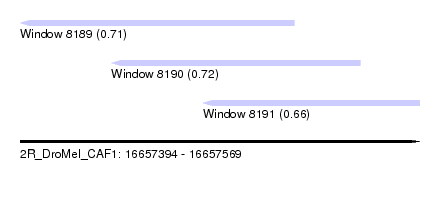
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,657,394 – 16,657,569 |
| Length | 175 |
| Max. P | 0.724687 |

| Location | 16,657,394 – 16,657,514 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -24.63 |
| Energy contribution | -26.05 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16657394 120 - 20766785 CAAGGAGGAUGAGGUCAUCGAUCUCAAGGCAAAGAUUGCCCAACUUUUGGCCGUGAUGCCAGACAACUUGUGCAUGAACACUGGCCCACAUGGGCCGUCGAGCAGCAUUCUGCGCAUGAA ((((..(..(((((((...))))))).((((.....)))).....((((((......)))))))..))))..((((......(((((....))))).....((((....)))).)))).. ( -39.30) >DroVir_CAF1 6863 120 - 1 CAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCACAGCUGCUGGCUGUUAUGCCGGAUACACUGUGUCUGAACACGGGAGCACAUGGUCCCGGCGGAAGUAUGCUGCGCAUGAA ......((.(((((((...)))))))...))......((.((((((((..((((.....(((((((...)))))))....(((((.(....).))))))))).)))).)))).))..... ( -43.10) >DroGri_CAF1 7271 120 - 1 CCAGGAUGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCACAGUUGCUGGCAGUUAUGCCAGACUCAAUGUGCCUAAAUACAGGACCACAUGGUCCAGGCGGUAGCAUGCUGCGCAUGAA .(((.(((.(((((((...)))))))..(((......(((((...(((((((....)))))).)....))))).........(((((....))))).))).....))).)))........ ( -44.90) >DroWil_CAF1 6317 111 - 1 UAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAAAUUGCCCAAUUGUUGGCUGUAAUGCCCGACAGCAUGUGCAUGAACACCGGUUC---------AGCUAGUAGCAUGUUGCGCAUGAA ...(..((.(((((((...))))))).((((((((((....)))).)))))).......))..)..(((((((((((((....))))---------)((.....))....)))))))).. ( -35.50) >DroMoj_CAF1 6714 117 - 1 CAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCCCAGCUGCUGGCUGUAAUGCCGGACACGCUGUGCCUAAACACGGGGCCGCAUGGU---AGCGGAAGCAUGCUGCGCAUGAA .........(((((((...))))))).((((......((.(((((((((((......))))).)).)))).))((......)))))).((((((---((((......)))))).)))).. ( -44.70) >DroAna_CAF1 5994 120 - 1 CAAGGAGGAAGAGGUCGUUGAUCUCAAGGCCAAGAUAGCGCAGCUACUGGCUGUAAUGCCGGACACACUGUGCAUGAACACGGGACCCCAUGGCCCGACCAGCAGCAUUCUGCGCAUGAA ............((((((((....)))(((((.....((((((...(((((......))))).....))))))........((....)).)))))))))).((((....))))....... ( -41.50) >consensus CAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCCCAGCUGCUGGCUGUAAUGCCGGACACACUGUGCAUGAACACGGGACCACAUGGUCCAGCCAGAAGCAUGCUGCGCAUGAA ...((....(((((((...)))))))...))......((.((((..(((((......))))).......((((.((......(((((....)))))......)))))))))).))..... (-24.63 = -26.05 + 1.42)
| Location | 16,657,434 – 16,657,543 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.66 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.57 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16657434 109 - 20766785 -----------AUAUCCACUCCCGCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUCGAUCUCAAGGCAAAGAUUGCCCAACUUUUGGCCGUGAUGCCAGACAACUUGUGCAUGAACA -----------............(((((((.(((((.(((....))))))))))))...........((((.....)))).....((((((......)))))).......)))....... ( -30.80) >DroVir_CAF1 6903 109 - 1 -----------UAUUUCUUGCUUCCAGGUCAUCGUCGGUCCAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCACAGCUGCUGGCUGUUAUGCCGGAUACACUGUGUCUGAACA -----------....((((((((..((((((.(.(((.(((.....)))))).)....))))))..))).)))))((((((((...(((((......))))).....))))).))).... ( -35.10) >DroPse_CAF1 8189 119 - 1 CUAAUAAC-GAUUACGCUUUCAUUCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUCGAUCUCAAGGCCAAGAUUGCCCAACUUCUGGCCGUGAUGCCAGACACAAUGUGCAUGAACA ........-.....(((.........((((.(((((.(((....))))))))))))...........(((((.((..........))))))))))((((((.......)).))))..... ( -32.70) >DroWil_CAF1 6348 109 - 1 -----------UUUUUCUUUUUUUAAGGUAAUCGCCGCUCUAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAAAUUGCCCAAUUGUUGGCUGUAAUGCCCGACAGCAUGUGCAUGAACA -----------...............((((((.(((.(((....)))..(((((((...))))))).)))....))))))....(((((((......).))))))((....))....... ( -27.30) >DroMoj_CAF1 6751 109 - 1 -----------CUGUUUUGGCUUGCAGGUCAACGUCGGUCCAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCCCAGCUGCUGGCUGUAAUGCCGGACACGCUGUGCCUAAACA -----------.(((((((((((..((((((((.(((.(((.....)))))).))...))))))..)))))))))))((.(((((((((((......))))).)).)))).))....... ( -44.20) >DroAna_CAF1 6034 116 - 1 CUAAUGGUUCCUUGUC----CUGGCAGACCAUCGUCGCUCCAAGGAGGAAGAGGUCGUUGAUCUCAAGGCCAAGAUAGCGCAGCUACUGGCUGUAAUGCCGGACACACUGUGCAUGAACA ......((((..((((----.((((.((((.((.((.(((....))))).)))))).(((....))).)))).))))((((((...(((((......))))).....))))))..)))). ( -43.50) >consensus ___________UUAUUCUUUCUUGCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCCCAACUGCUGGCUGUAAUGCCGGACACACUGUGCAUGAACA .................................(((..(((.....)))(((((((...))))))).))).......((.(((...(((((......))))).....))).))....... (-20.87 = -20.57 + -0.30)

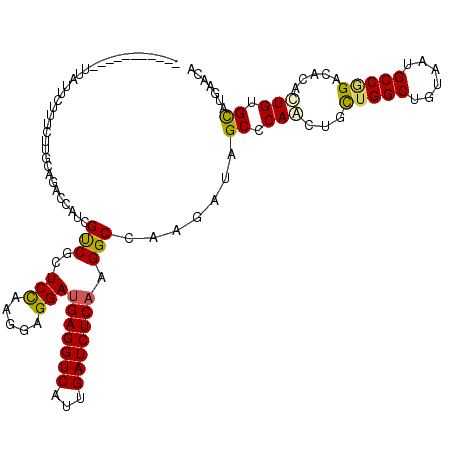
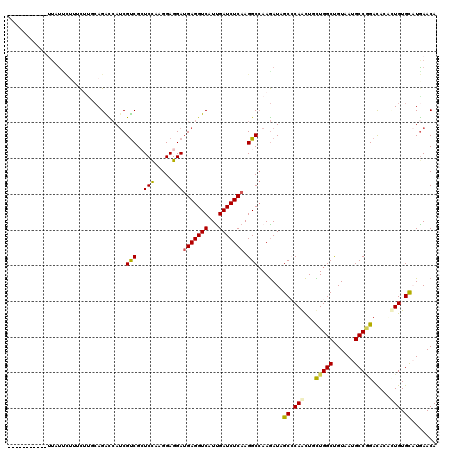
| Location | 16,657,474 – 16,657,569 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.30 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16657474 95 - 20766785 UGUUUUAAUG--U-UGAUAGCUAGUUGAU----------AUAUCCACUCCCGCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUCGAUCUCAAGGCAAAGAUUGCC ((((((.(.(--(-((((.((.(((.((.----------...)).)))...)).((((.(((((.(((....)))))))))))))))))).).))))))......... ( -26.20) >DroVir_CAF1 6943 92 - 1 --UCAGAAUGAUUGUAAUAAACA----AC----------UAUUUCUUGCUUCCAGGUCAUCGUCGGUCCAAGGAGGAUGAGGUCAUUGAUCUCAAGGCCAAGAUAGCA --...(((((.((((.....)))----).----------)))))..((((((..((((.......((((.....))))((((((...))))))..))))..)).)))) ( -22.20) >DroSec_CAF1 5983 105 - 1 CAUUUAAAUG--U-UGAUAGCUAGUUGAUUUCUAAUGCAAUAUCCACUCACGCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUCGAUCUCAAGGCAAAGAUUGCC .........(--(-((((.((.(((.(((............))).)))...)).((((.(((((.(((....)))))))))))))))))).....((((.....)))) ( -26.70) >DroSim_CAF1 6008 105 - 1 AGUUUAAAUG--U-UGAUAGCUAGUUGAUUUCUAAUGCAAUAUCCACUCACGCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUCGAUCUCAAGGCAAAGAUUGCC .........(--(-((((.((.(((.(((............))).)))...)).((((.(((((.(((....)))))))))))))))))).....((((.....)))) ( -26.70) >DroEre_CAF1 6053 102 - 1 -GU-UUAAUG--U-A-AAAGCUGGUUGAUUUCUAAUUCAUUAUUCACUUCCUCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUUGAUCUCAAGGCAAAGAUUGCC -..-....((--.-.-.(((.(((.(((........)))....))))))...))((((.(((((.(((....)))))))))))).(((....)))((((.....)))) ( -23.20) >DroYak_CAF1 6034 101 - 1 -GU-UUAAUG--U-A-AAAGA-AGUUGAUUUUAAAUUCAUUAUUUACUUCCUCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUUGAUCUCAAGGCAAAGAUUGCC -..-(((((.--.-.-...((-(((.(((............))).)))))....((((.(((((.(((....)))))))))))))))))......((((.....)))) ( -24.60) >consensus _GUUUUAAUG__U_UGAUAGCUAGUUGAUUUCUAAU_CAAUAUCCACUCCCGCAGACCAUCGUCGCUCCAAGGAGGAUGAGGUCAUCGAUCUCAAGGCAAAGAUUGCC ......................................................((((.(((((.(((....))))))))))))..((((((........)))))).. (-17.49 = -17.30 + -0.19)

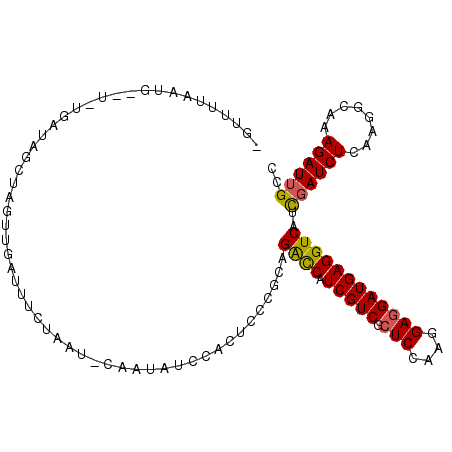
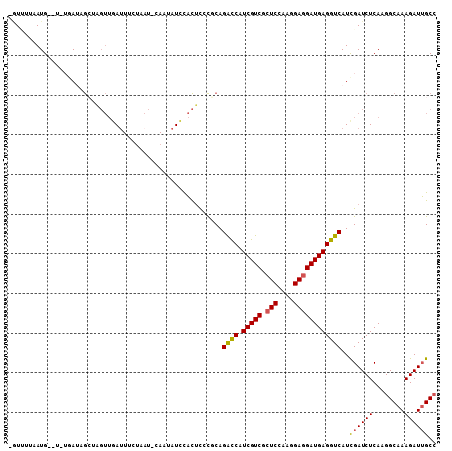
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:06 2006