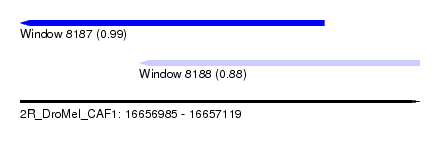
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,656,985 – 16,657,119 |
| Length | 134 |
| Max. P | 0.988741 |

| Location | 16,656,985 – 16,657,087 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
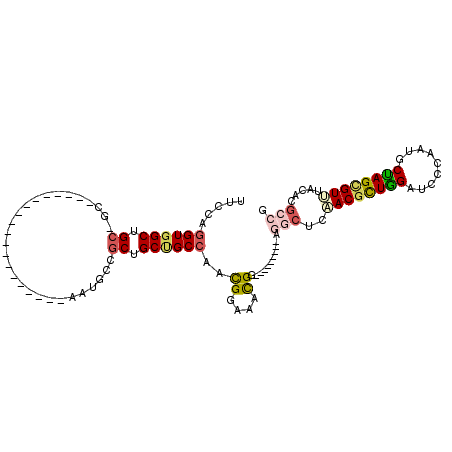
>2R_DroMel_CAF1 16656985 102 - 20766785 UUCCAGGUGGCUGCAGCAAAUGCCGCUGCUGCAGCCAAUGCCGCUGCUGCCAACGGAAACGG---------GGGCUCAACGCUGGAUCCCAAUGCUAGCGUUUACACGCCG .(((((.(((((((((((........)))))))))))....((.((..(((..((....)).---------.))).)).)))))))...........((((....)))).. ( -45.20) >DroPse_CAF1 7719 93 - 1 CUCCAGGUGGCUGCUGCC------------------AAUGCAGCUGCCGCAAAUGGGAACGGCAACAAUGGCAACUCCACACUGGAUCCCAAUGCCAGCGUCUACACGCCC .((((.(((((.(((((.------------------...))))).)))))...))))...((((....(((....(((.....)))..))).)))).((((....)))).. ( -35.70) >DroSec_CAF1 5518 84 - 1 UUCCAGGUGGCUGC------------------AGCCAACGCCGCUGCUGCCAACGGAAACGG---------AGGCUCAACGCUAGAUCCCAAUGCUAGCGUUUACACGCCG .(((.(.((((.((------------------(((.......))))).)))).)(....)))---------)(((..((((((((.........)))))))).....))). ( -34.10) >DroSim_CAF1 5537 84 - 1 UUCCAGGUGGCUGC------------------AGCCAAUGCCGCUGCUGCCAACGGAAACGG---------AGGCUCAACGCUGGAUCCCAAUGCUAGCGUUUACACGCCG .(((.(.((((.((------------------(((.......))))).)))).)(....)))---------)(((..((((((((.........)))))))).....))). ( -33.80) >DroYak_CAF1 5569 87 - 1 UUCCAGGUUGCUGCCGCCAAUGCCAAUG---------------CUGCUGCCAACGGAAAUGG---------GGGCUCAACGCUGGAUCCCAAUGCUAGCGUGUACACGCCG .(((((((((..(((.(((...((...(---------------(....))....))...)))---------.))).)))).)))))...........((((....)))).. ( -30.60) >DroAna_CAF1 5627 84 - 1 CUGCAGGUGGCUGCGGCA------------------AAUGCUGCUGCCGCCAACGGAAACGG---------UUCGUCCACGUUGGAUCCUAACGCCAGUGUGUACACGCCC ..((.((((((.(((((.------------------...))))).))))))..((....)).---------...((.((((((((.........)))))))).))..)).. ( -35.50) >consensus UUCCAGGUGGCUGC_GC___________________AAUGCCGCUGCUGCCAACGGAAACGG_________AGGCUCAACGCUGGAUCCCAAUGCUAGCGUUUACACGCCG .....((((((.((............................)).))))))..((....))...........(((..((((((((.........)))))))).....))). (-18.56 = -19.17 + 0.62)

| Location | 16,657,025 – 16,657,119 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
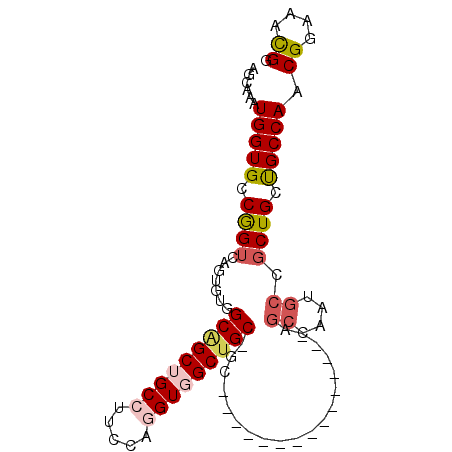
>2R_DroMel_CAF1 16657025 94 - 20766785 AGCAAAUGGUGCCGGUCAGUGUGGCAGCUGCCUUCCAGGUGGCUGCAGCAAAUGCCGCUGCUGCAGCCAAUGCCGCUGCUGCCAACGGAAACGG .(((.....))).((.((((((((((...(((.....)))((((((((((........))))))))))..)))))).))))))..((....)). ( -46.00) >DroSec_CAF1 5558 76 - 1 AGCAAAUGGUGCCGGUCAGUGUGGCGGCUGCCUUCCAGGUGGCUGC------------------AGCCAACGCCGCUGCUGCCAACGGAAACGG .(((.....))).((.(((((((((((((((...((....))..))------------------))))...))))).))))))..((....)). ( -33.30) >DroSim_CAF1 5577 76 - 1 AGCAAAUGGUGCCGGUCAGUGUGGCGGCUGCCUUCCAGGUGGCUGC------------------AGCCAAUGCCGCUGCUGCCAACGGAAACGG .(((.....))).((.(((((((((((((((...((....))..))------------------))))...))))).))))))..((....)). ( -33.30) >DroEre_CAF1 5613 94 - 1 AGCAAAUGGUGCCGGUCAGUGUCGCAGCUGCCUUCCAAGUUGCUGCAGCCAAUGCCGCUGCUGCAGCCAAUGCCGCUGCUGCCAACGGAAACGG .(((...((..(......)..))(((((.((.......(((((.(((((.......))))).)))))....)).))))))))...((....)). ( -34.20) >DroYak_CAF1 5609 79 - 1 AGCAAAUGGUGCCGGUCAGUGUGGCAGCUGCCUUCCAGGUUGCUGCCGCCAAUGCCAAUG---------------CUGCUGCCAACGGAAAUGG .(((.....))).((.((((((((((((.(((.....))).))))))))....((....)---------------).))))))........... ( -28.20) >DroAna_CAF1 5667 76 - 1 AGCAGAUGGUGCCAGUCAGUGUGGCUGCUGCACUGCAGGUGGCUGCGGCA------------------AAUGCUGCUGCCGCCAACGGAAACGG .((((...((..(((((.....)))))..)).)))).((((((.(((((.------------------...))))).))))))..((....)). ( -39.40) >consensus AGCAAAUGGUGCCGGUCAGUGUGGCAGCUGCCUUCCAGGUGGCUGC_GC_______________AGCCAAUGCCGCUGCUGCCAACGGAAACGG ......(((((.((((.......(((((((((.....)))))))))...................((....)).)))).))))).((....)). (-20.92 = -21.75 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:03 2006