
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,783,274 – 2,783,377 |
| Length | 103 |
| Max. P | 0.699364 |

| Location | 2,783,274 – 2,783,377 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.19 |
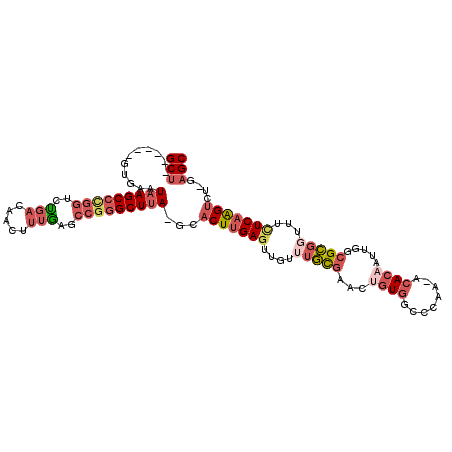
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -23.83 |
| Energy contribution | -25.95 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
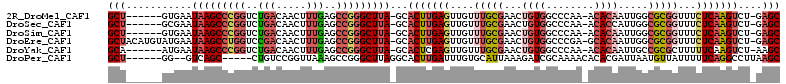
>2R_DroMel_CAF1 2783274 103 + 20766785 GCU------GUGAAUAAGCCCGGUCUGACAACUUUGAGCCGGGCUUA-GCACUUGAGUUGUUUGCGAACUGUGGCCCAA-ACACAAUUGGCGCGGUUUCUCAAGUCU-GAGC (((------.....((((((((((..((.....))..))))))))))-(.(((((((....(((((...((((......-.)))).....)))))...)))))))).-.))) ( -37.20) >DroSec_CAF1 17250 103 + 1 GCU------GCGAAUAAGCCCGGUCUGACAACUUUGAGCCGGGCUUA-GCACUUGAGUUGUUUGCGAACUGUGGCCCAA-ACACCAUUGGCGCGGUUUCUCAAGUCU-GAGC (((------((...((((((((((..((.....))..))))))))))-))(((((((....(((((....((((.....-...))))...)))))...)))))))..-.))) ( -38.60) >DroSim_CAF1 31547 103 + 1 GCU------GUGAAUAAGCCCGGUCUGACAACUUUGAGCCGGGCUUA-GCACUUGAGUUGUUUGCGAACUGUGGCCCAA-ACACAAUUGGCGCGGUUUCUCAAGUCU-GAGC (((------.....((((((((((..((.....))..))))))))))-(.(((((((....(((((...((((......-.)))).....)))))...)))))))).-.))) ( -37.20) >DroEre_CAF1 14645 109 + 1 GCUACAUGUAUGAAUAAGCCUGGUCCGACAACUUUGAGCCGGGCUUA-GCACUUGAGUUGUUUGCGAACUGUGGCCCGA-GCACAAUUGGCGCGGUUUCUCAAGUCU-GAGC (((.((........(((((((((..(((.....)))..)))))))))-(.(((((((........((((((((..((((-......)))))))))))))))))))))-)))) ( -36.80) >DroYak_CAF1 14695 103 + 1 GCA------AUGAAUAAGCCCGGUCUGACAACUUUGAGCCGGGCUUA-GCACUCGAGUUGUUUGCGAACUGUGGCCCAA-ACACAAUUGCCGCGCUUUUUCAAGUCU-AAGC ...------.((((((((((((((..((.....))..))))))))))-((.....(((((....).))))(((((....-........)))))))...)))).....-.... ( -29.20) >DroPer_CAF1 15485 99 + 1 GCU------GG--GUCAGC-----CUGUCCGGUUAAAGCCGGGCUUAGGCACUUGAUUUGUGCAUUAAAGAUCGCAAAACACACGAUUAAUGUUAUUUUUCAGGCCUUAAGC (((------((--(((.((-----(((((((((....)))))))..))))...(((.....((((((...((((.........))))))))))......))))))))..))) ( -32.50) >consensus GCU______GUGAAUAAGCCCGGUCUGACAACUUUGAGCCGGGCUUA_GCACUUGAGUUGUUUGCGAACUGUGGCCCAA_ACACAAUUGGCGCGGUUUCUCAAGUCU_GAGC (((...........(((((((((..(((.....)))..)))))))))...(((((((....(((((...((((........)))).....)))))...)))))))....))) (-23.83 = -25.95 + 2.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:35 2006