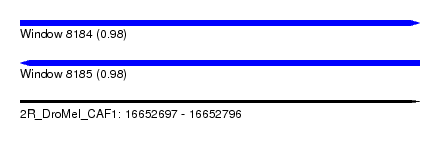
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,652,697 – 16,652,796 |
| Length | 99 |
| Max. P | 0.979674 |

| Location | 16,652,697 – 16,652,796 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -30.58 |
| Energy contribution | -29.36 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16652697 99 + 20766785 UACUGUGACGGCGUCAGCGCUGGCGACG------ACUCAACGGACGUGAUCAUGAAGUGUCCGCCGCCAGCGACGUCUCCGUCACAGGUCUAUCUCAACGUGAUG ..(((((((((.(.(..(((((((((..------..))..((((((...........))))))..)))))))..).).)))))))))...((((.......)))) ( -40.50) >DroVir_CAF1 1301 99 + 1 UACGGCGAGGGCGUCAGUGCUGGCGA------GGACUCUACAGAUGUUAUCAUGAAGUGCCCGCCGCCAGCAACGUCGCCUUCGCAGGUUUACCUCAACGUGAUA (((((((((((((.(..((((((((.------((.(.((.(((((...))).)).)).).))..))))))))..).)))))))))(((....)))...))))... ( -46.10) >DroGri_CAF1 1650 105 + 1 UAUGGCGAGGGUGUCACUGCUGGCGAUGAGGAGAACACAACGGAUGCCAUCAUGAAGUGUCCGCUGCCAGCGACGUCACCCUCCCAGGUUUAUCUCAACGUCAUC ..(((.(((((((.(..((((((((.((.(.....).)).(((((((.........))))))).))))))))..).))))))))))................... ( -38.60) >DroMoj_CAF1 1321 102 + 1 UACGGUGAGGGCGUCACUGCUGGCGAUGA---GAACACCACAGAUGCGAUCAUGAAGUGCCCGUUGCCAGCAACGUCGCCCUCACAGGUCUACCUCAAUGUGAUC (((((((((((((.(..(((((((((((.---(..(((..(((((...))).))..))).))))))))))))..).)))))))))(((....)))...))))... ( -43.70) >DroAna_CAF1 1271 99 + 1 UACAGCGACGGUGUCACCGCUGGCGACG------ACGCCACAGAUGUGGUGAUGAAAUGUCCGCCGCCAGCGACGUCGCCGUCGCAAGUCUAUCUCAAUGUGGUG ....(((((((((.(..((((((((.((------.((((((....))))))..((....)))).))))))))..).))))))))).................... ( -46.60) >DroPer_CAF1 2640 99 + 1 UACUGCGAUGGCGUCAGUGCUGGGGAUG------ACACUACAGAUGUAAUUAUGAAGUGUCCGCCGCCAGCAACGUCCCCAUCGCAGGUCUAUCUCAACGUCGUG ..(((((((((.(.(..((((((((..(------(((((.((..........)).))))))..)).))))))..).).))))))))).................. ( -41.20) >consensus UACGGCGAGGGCGUCACUGCUGGCGAUG______ACACAACAGAUGUGAUCAUGAAGUGUCCGCCGCCAGCAACGUCGCCCUCGCAGGUCUAUCUCAACGUGAUG ....(((((((((.(..((((((((.......................................))))))))..).))))))))).................... (-30.58 = -29.36 + -1.22)

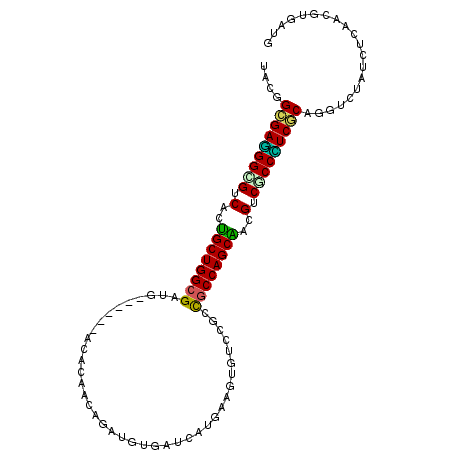
| Location | 16,652,697 – 16,652,796 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -43.18 |
| Consensus MFE | -28.05 |
| Energy contribution | -27.36 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16652697 99 - 20766785 CAUCACGUUGAGAUAGACCUGUGACGGAGACGUCGCUGGCGGCGGACACUUCAUGAUCACGUCCGUUGAGU------CGUCGCCAGCGCUGACGCCGUCACAGUA ..................(((((((((.(.((.(((((((((((...((((.(((........))).))))------))))))))))).)).).))))))))).. ( -43.70) >DroVir_CAF1 1301 99 - 1 UAUCACGUUGAGGUAAACCUGCGAAGGCGACGUUGCUGGCGGCGGGCACUUCAUGAUAACAUCUGUAGAGUCC------UCGCCAGCACUGACGCCCUCGCCGUA ....(((...(((....)))((((.((((.((.(((((((((.((((.((.((.(((...))))).)).))))------))))))))).)).)))).))))))). ( -43.80) >DroGri_CAF1 1650 105 - 1 GAUGACGUUGAGAUAAACCUGGGAGGGUGACGUCGCUGGCAGCGGACACUUCAUGAUGGCAUCCGUUGUGUUCUCCUCAUCGCCAGCAGUGACACCCUCGCCAUA ......(((......))).((((((((((.((..((((((.(.(((.....((..((((...))))..))...))).)...))))))..)).))))))).))).. ( -40.30) >DroMoj_CAF1 1321 102 - 1 GAUCACAUUGAGGUAGACCUGUGAGGGCGACGUUGCUGGCAACGGGCACUUCAUGAUCGCAUCUGUGGUGUUC---UCAUCGCCAGCAGUGACGCCCUCACCGUA ....((....(((....)))(((((((((.((((((((((...(((((((.((.(((...))))).)))))))---.....)))))))))).))))))))).)). ( -44.90) >DroAna_CAF1 1271 99 - 1 CACCACAUUGAGAUAGACUUGCGACGGCGACGUCGCUGGCGGCGGACAUUUCAUCACCACAUCUGUGGCGU------CGUCGCCAGCGGUGACACCGUCGCUGUA ................((..(((((((.(.(..((((((((((((.........(.((((....)))).))------)))))))))))..).).))))))).)). ( -45.60) >DroPer_CAF1 2640 99 - 1 CACGACGUUGAGAUAGACCUGCGAUGGGGACGUUGCUGGCGGCGGACACUUCAUAAUUACAUCUGUAGUGU------CAUCCCCAGCACUGACGCCAUCGCAGUA ..................(((((((((.(.((.((((((.((..((((((.((..........)).)))))------)..)))))))).)).).))))))))).. ( -40.80) >consensus CAUCACGUUGAGAUAGACCUGCGACGGCGACGUCGCUGGCGGCGGACACUUCAUGAUCACAUCUGUAGUGU______CAUCGCCAGCACUGACGCCCUCGCCGUA ....................(((((((.(.((.(((((((((((((...............)))))..............)))))))).)).).))))))).... (-28.05 = -27.36 + -0.69)

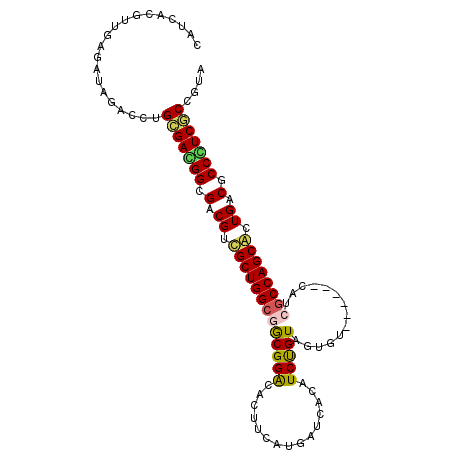

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:00 2006