
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,636,686 – 16,636,786 |
| Length | 100 |
| Max. P | 0.895009 |

| Location | 16,636,686 – 16,636,786 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.29 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.895009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16636686 100 + 20766785 GGGCCUGCGAUGCUUUUACUACCUGGUGCAGGAUCUCAAGUGCCUGGUCUUCUCACUCAUCGGCCUGCAUUUCAAGAUCAAGCCCAUAUAAG--CCGUAG----------CC-- .(((.((((..((((.((.....(((.((..(((((.((((((..((((............)))).))))))..)))))..))))))).)))--))))))----------))-- ( -30.70) >DroPse_CAF1 41118 108 + 1 GGGCUUGCGCUGCUUCUAUUAUCUGGUGCAAGAUCUCAAGUGUUUGGUCUUCUCGCUCAUCGGCCUGCACUUCAAGAUCAAGCCCAUAUAAG--CUGCCC-ACUCUCAU-AU-- (((((((((((.............)))))))(((((.((((((..((((............)))).))))))..))))).(((........)--))))))-........-..-- ( -32.02) >DroWil_CAF1 57085 109 + 1 AGGCUUGAGAUGCUUCUAUUAUCUAGUGCAGGAUCUUAAGUGUUUGGUCUUCUCACUCAUCGGUUUGCACUUCAAGAUCAAGCCCAUAUAGA--AGGCUUCUCUCACGU-AU-- ..((.(((((.((((((((......(.((..(((((((((((((((((..........)))))...)))))).))))))..)))...)))))--).))...))))).))-..-- ( -26.90) >DroMoj_CAF1 53741 102 + 1 AGGCUUGCGCUGCUUCUAUUAUUUGGUACAGGAUUUAAAGUGUCUGGUCUUCUCGCUCAUCGGUCUCCACUUCAAAAUUAAACCCAUAUAAAACU------------UCUCCGA .(((....))).......((((.(((.....(((((.(((((.(((((..........)))))....)))))..)))))....))).))))....------------....... ( -12.40) >DroAna_CAF1 39525 99 + 1 GGGACUGCGCUGCUUCUACUAUUUGGUGCAGGAUCUCAAGUGUUUGGUCUUCUCACUCAUUGGCCUGCAUUUCAAGAUCAAACCCAUAUAAGAACCG-------------AU-- (((.(((((((.............)))))))(((((.((((((..((((............)))).))))))..)))))...)))............-------------..-- ( -28.72) >DroPer_CAF1 41681 108 + 1 GGGCUUGCGCUGCUUCUAUUAUCUGGUGCAAGAUCUCAAGUGUUUGGUCUUCUCGCUCAUCGGCCUGCACUUCAAGAUCAAGCCCAUAUAAG--CUGCCC-ACUCUCAU-AU-- (((((((((((.............)))))))(((((.((((((..((((............)))).))))))..))))).(((........)--))))))-........-..-- ( -32.02) >consensus GGGCUUGCGCUGCUUCUAUUAUCUGGUGCAGGAUCUCAAGUGUUUGGUCUUCUCACUCAUCGGCCUGCACUUCAAGAUCAAGCCCAUAUAAG__CCGC__________U_AU__ (((((((((((.............)))))))(((((.((((((..((((............)))).))))))..)))))..))))............................. (-20.48 = -20.29 + -0.19)
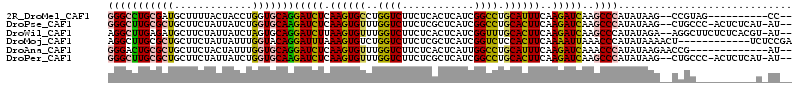

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:50 2006