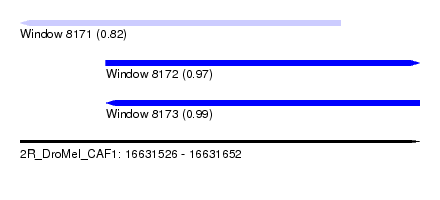
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,631,526 – 16,631,652 |
| Length | 126 |
| Max. P | 0.994017 |

| Location | 16,631,526 – 16,631,627 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.06 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16631526 101 - 20766785 GUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGAUAUGUGGUCCAUUCCACACAAUUCCGUUGUACUUAUUAGUUAUUAA ...........((((((((((.(((((((.((..........(((...)))......(((((......))))))))))))))))).)))).)))....... ( -20.60) >DroSec_CAF1 30858 99 - 1 GUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGUCCAUUCCACACAAUUCCGUUGUACUUAUUAGUUAUUAA ...........((((((((((.(((((((.((..........(((...)))....--(((((......))))))))))))))))).)))).)))....... ( -20.60) >DroSim_CAF1 30746 99 - 1 GUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGUCCAUUCCACACAAUUCCGUUGUACUUAUUAGUUAUUAA ...........((((((((((.(((((((.((..........(((...)))....--(((((......))))))))))))))))).)))).)))....... ( -20.60) >DroEre_CAF1 33900 99 - 1 GUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGAGUAUUCCACGCAAUUCCGUUGUACUUAUUAGUUAUUAA ...........((((((((((.(((((((.((..........(((...)))....--.((((......)))).)))))))))))).)))).)))....... ( -20.20) >DroYak_CAF1 32196 99 - 1 GUUCUCACAUUCUGUGAGACAAACAGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGAGCAUUCCACGCAAUUCCGUUGUACUUAUUAGUUAAUAA ..((((((.....))))))..........(((((((..(((((.....(.(((..--(((((......)))))...)))))))))..)))))))....... ( -20.80) >consensus GUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA__UGUGGUCCAUUCCACACAAUUCCGUUGUACUUAUUAGUUAUUAA ...........((((((((((.(((((((.((..........(((...)))......(((((......))))))))))))))))).)))).)))....... (-19.10 = -19.06 + -0.04)

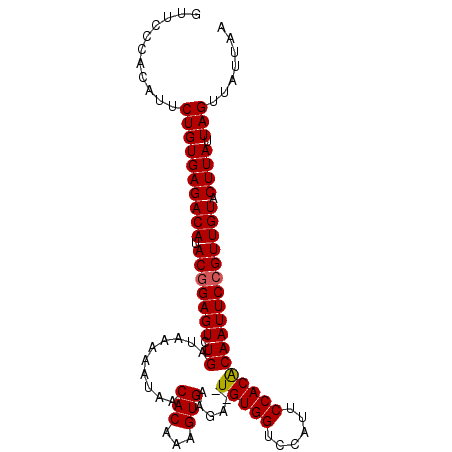
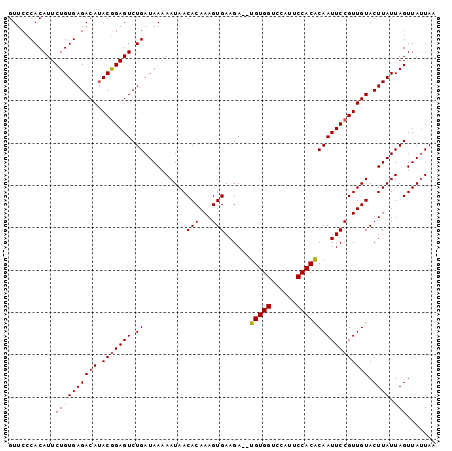
| Location | 16,631,553 – 16,631,652 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -18.94 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16631553 99 + 20766785 GUGUGGAAUGGACCACAUAUCUUCACUUUGUGUUAUUUUUAUCAGACUCCGUAUGUCUCACAGAAUGUGGGAACCUUGCGUUCCACUAACUCAACUCUU ..(((((((((.((((((........((((((...........((((.......))))))))))))))))...))....)))))))............. ( -21.80) >DroSec_CAF1 30885 97 + 1 GUGUGGAAUGGACCACA--UCUUCACUUUGUGUUAUUUUUAUCAGACUCCGUAUGUCUCACAGAAUGUGGGAACCUUGCGUUCCACCAACUCAAAUCUU (.(((((((((.(((((--(......((((((...........((((.......))))))))))))))))...))....))))))))............ ( -22.10) >DroSim_CAF1 30773 97 + 1 GUGUGGAAUGGACCACA--UCUUCACUUUGUGUUAUUUUUAUCAGACUCCGUAUGUCUCACAGAAUGUGGGAACCUUGCGUUCCACCAACUCAACUCUU (.(((((((((.(((((--(......((((((...........((((.......))))))))))))))))...))....))))))))............ ( -22.10) >DroEre_CAF1 33927 97 + 1 GCGUGGAAUACUCCACA--UCUUCACUUUGUGUUAUUUUUAUCAGACUCCGUAUGUCUCACAGAAUGUGGGAACCUUUAGUUCCACUAACUCAACUUUU ..(((((....))))).--....(((((((((...........((((.......))))))))))..)))(((((.....)))))............... ( -20.80) >DroYak_CAF1 32223 97 + 1 GCGUGGAAUGCUCCACA--UCUUCACUUUGUGUUAUUUUUAUCAGACUCUGUUUGUCUCACAGAAUGUGAGAACCUUUCGUUCCACUAACUCAACUUUU ..((((((((...((((--.........))))..........(((((...)))))((((((.....))))))......))))))))............. ( -21.80) >consensus GUGUGGAAUGGACCACA__UCUUCACUUUGUGUUAUUUUUAUCAGACUCCGUAUGUCUCACAGAAUGUGGGAACCUUGCGUUCCACUAACUCAACUCUU ..((((((((...((((...........)))).......................((((((.....))))))......))))))))............. (-18.94 = -18.98 + 0.04)

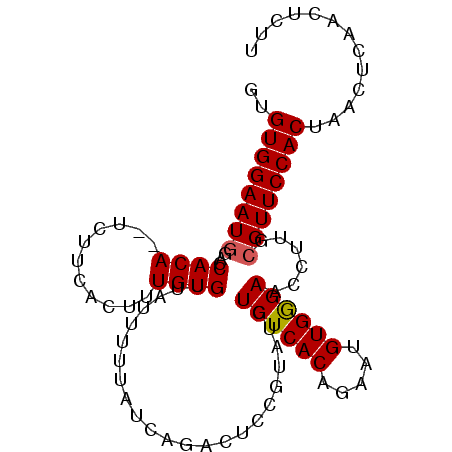
| Location | 16,631,553 – 16,631,652 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -22.34 |
| Energy contribution | -21.86 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16631553 99 - 20766785 AAGAGUUGAGUUAGUGGAACGCAAGGUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGAUAUGUGGUCCAUUCCACAC .............(((((((....)....(((((((((((.((((.......)))).))........(((...))).))).))))))....)))))).. ( -23.80) >DroSec_CAF1 30885 97 - 1 AAGAUUUGAGUUGGUGGAACGCAAGGUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGUCCAUUCCACAC .............(((((((....)....(((((((.(((.((((.......)))).))).......(((...)))..))--)))))....)))))).. ( -23.70) >DroSim_CAF1 30773 97 - 1 AAGAGUUGAGUUGGUGGAACGCAAGGUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGUCCAUUCCACAC .............(((((((....)....(((((((.(((.((((.......)))).))).......(((...)))..))--)))))....)))))).. ( -23.70) >DroEre_CAF1 33927 97 - 1 AAAAGUUGAGUUAGUGGAACUAAAGGUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGAGUAUUCCACGC .....(((.((((..(((((.....)))))((.((((((((.....)))))))).)).......)))).)))........--.((((......)))).. ( -22.40) >DroYak_CAF1 32223 97 - 1 AAAAGUUGAGUUAGUGGAACGAAAGGUUCUCACAUUCUGUGAGACAAACAGAGUCUGAUAAAAAUAACACAAAGUGAAGA--UGUGGAGCAUUCCACGC .............(((((((....)((((.((((((.(((.((((.......)))).))).......(((...)))..))--)))))))).)))))).. ( -26.40) >consensus AAGAGUUGAGUUAGUGGAACGCAAGGUUCCCACAUUCUGUGAGACAUACGGAGUCUGAUAAAAAUAACACAAAGUGAAGA__UGUGGUCCAUUCCACAC .....(((.(((((((((((.....)))).)))....(((.((((.......)))).)))....)))).)))..........(((((......))))). (-22.34 = -21.86 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:49 2006