
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,613,494 – 16,613,594 |
| Length | 100 |
| Max. P | 0.975222 |

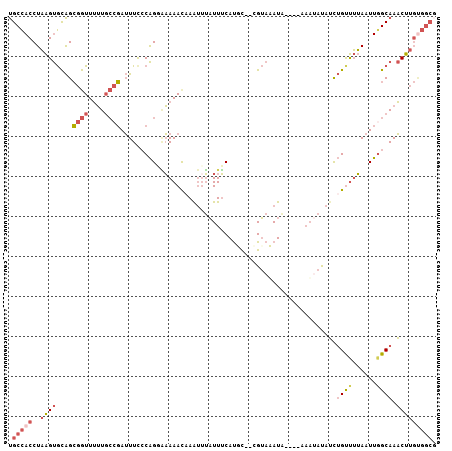
| Location | 16,613,494 – 16,613,594 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -9.36 |
| Energy contribution | -10.78 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16613494 100 + 20766785 UGCCACCUAAGUGCAGCGGUUUUUGCCGAUUUUCCAGUAAAAACAAAUUUAUUUCAUGC--CGUAUAUG----A-AUAUAUCUGUUUUAAUUGGCAAACUUGUGGCG .(((((.....(((..((((....))))......((((.((((((.((....((((((.--....))))----)-)...)).)))))).))))))).....))))). ( -27.60) >DroSec_CAF1 13421 101 + 1 UGCCACCUAAGUGCAGCGGUUUUUGCCGAUUUCCCAGGAAAAACAAAUUUAUUUCAUGC--CGUAUAUG----AAAUAUAUCUGUUUUAAUUGGCAAACUUGUGGCG .(((((.....(((..((((....))))......(((..((((((.((.(((((((((.--....))))----))))).)).))))))..)))))).....))))). ( -30.90) >DroSim_CAF1 13001 103 + 1 UGCCACCUAAGUGCAGCGGUUUUUGCCGAUUUCCCAGGAAAAACAAAUUUAUUUCAUGCCCCGUAUAUG----AAAUAUAUCUGUUUUAAUUGGCAAACUUGUGGCG .(((((.....(((..((((....))))......(((..((((((.((.(((((((((.......))))----))))).)).))))))..)))))).....))))). ( -30.60) >DroEre_CAF1 15754 99 + 1 UGCCACCUAAGUGCAGCGGUUUUU-CCGAUUUCCCAGGAAAAACCAGUUUAUUUCAUUC--CGUAAAUA-----AAUACAUCUGCUUUAAUUGGCAAACUUGUGGCG .(((((.....(((((((((((((-((.........))))))))).(((((((((....--.).)))))-----)))......))).......))).....))))). ( -28.61) >DroYak_CAF1 13685 100 + 1 UGCCACCUAAGUGCAGCGGUUUUUGCCGAUUUUCCAGCGAAAAUAAAUGUAUUUCAUUU--CGUAAAUA-----AUCACAUCUGUUUUAAUUUGCAAACUUGCGGCU .(((....(((((((...((((((((.(......).))))))))...))))))).....--.((((((.-----(..((....))..).))))))........))). ( -21.70) >DroAna_CAF1 15437 98 + 1 CGCUACCUUGGCAUGGAAAUUGGUGAAUAAUUCACGGCCUAAACAAACUGUUUGCACUC--AGAAAAUAAAUAAAACAUAAUGGCCAAAGUUGGCAAUCU------- .(((((.(((((..........((((.....)))).((..((((.....))))))....--......................))))).).)))).....------- ( -17.70) >consensus UGCCACCUAAGUGCAGCGGUUUUUGCCGAUUUCCCAGGAAAAACAAAUUUAUUUCAUGC__CGUAAAUA____AAAUAUAUCUGUUUUAAUUGGCAAACUUGUGGCG .(((((..((((....((((....))))......................................................((((......)))).))))))))). ( -9.36 = -10.78 + 1.42)


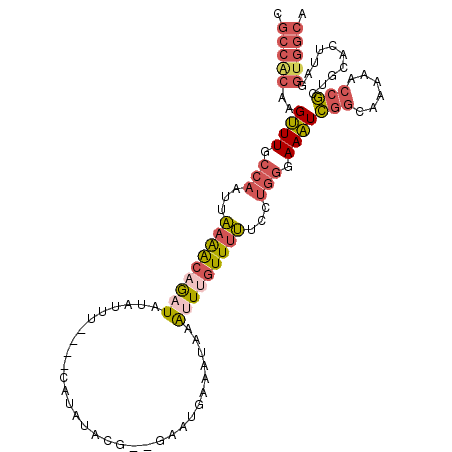
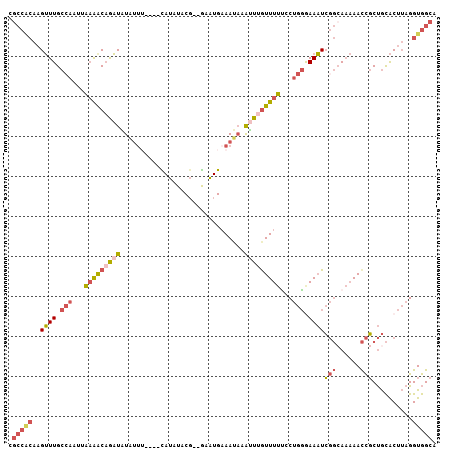
| Location | 16,613,494 – 16,613,594 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -10.79 |
| Energy contribution | -11.88 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16613494 100 - 20766785 CGCCACAAGUUUGCCAAUUAAAACAGAUAUAU-U----CAUAUACG--GCAUGAAAUAAAUUUGUUUUUACUGGAAAAUCGGCAAAAACCGCUGCACUUAGGUGGCA .(((((..((((.(((...(((((((((.(((-(----(((.....--..))).)))).)))))))))...))).))))((((.......)))).......))))). ( -25.60) >DroSec_CAF1 13421 101 - 1 CGCCACAAGUUUGCCAAUUAAAACAGAUAUAUUU----CAUAUACG--GCAUGAAAUAAAUUUGUUUUUCCUGGGAAAUCGGCAAAAACCGCUGCACUUAGGUGGCA .(((((..((((.(((...(((((((((.(((((----(((.....--..)))))))).)))))))))...))).))))((((.......)))).......))))). ( -32.40) >DroSim_CAF1 13001 103 - 1 CGCCACAAGUUUGCCAAUUAAAACAGAUAUAUUU----CAUAUACGGGGCAUGAAAUAAAUUUGUUUUUCCUGGGAAAUCGGCAAAAACCGCUGCACUUAGGUGGCA .(((((..((((.(((...(((((((((.(((((----(((.........)))))))).)))))))))...))).))))((((.......)))).......))))). ( -31.90) >DroEre_CAF1 15754 99 - 1 CGCCACAAGUUUGCCAAUUAAAGCAGAUGUAUU-----UAUUUACG--GAAUGAAAUAAACUGGUUUUUCCUGGGAAAUCGG-AAAAACCGCUGCACUUAGGUGGCA .(((((..((((((........))))))(((((-----(((((...--.....)))))))..(((((((((.((....))))-)))))))..)))......))))). ( -28.80) >DroYak_CAF1 13685 100 - 1 AGCCGCAAGUUUGCAAAUUAAAACAGAUGUGAU-----UAUUUACG--AAAUGAAAUACAUUUAUUUUCGCUGGAAAAUCGGCAAAAACCGCUGCACUUAGGUGGCA .(((((.....((((....((((.(((((((.(-----((((....--.)))))..))))))).)))).((((......)))).........)))).....))))). ( -22.70) >DroAna_CAF1 15437 98 - 1 -------AGAUUGCCAACUUUGGCCAUUAUGUUUUAUUUAUUUUCU--GAGUGCAAACAGUUUGUUUAGGCCGUGAAUUAUUCACCAAUUUCCAUGCCAAGGUAGCG -------.....((..((((((((((...(((((((((((.....)--))))).)))))...))........((((.....))))..........)))))))).)). ( -20.50) >consensus CGCCACAAGUUUGCCAAUUAAAACAGAUAUAUUU____CAUAUACG__GAAUGAAAUAAAUUUGUUUUUCCUGGGAAAUCGGCAAAAACCGCUGCACUUAGGUGGCA .(((((..((((.(((...(((((((((...............................)))))))))...))).))))(((......)))..........))))). (-10.79 = -11.88 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:43 2006