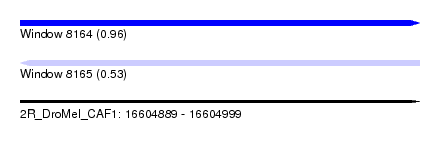
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,604,889 – 16,604,999 |
| Length | 110 |
| Max. P | 0.959336 |

| Location | 16,604,889 – 16,604,999 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -14.60 |
| Energy contribution | -16.64 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16604889 110 + 20766785 UUCAAAUUUCGUUAUCGACUUAUCGAUAGCCACGGCCAUGUGUUUCAGUAUAUACGGUUGUAUAUUAUACGGUCACACUAGGCACAAGGGGCGGAUUUUUUGAAACUUC----U ((((((..((((((((((....))))))))..((.((.(((((((.(((((((((....)))))).....(....)))))))))))..)).))))...)))))).....----. ( -29.00) >DroSec_CAF1 4852 111 + 1 UUCAAAUUUCGUUAUCGACCUAUCGAUAGCCAUGGCCAUGUGAUUCAGUAUAUACUUUUCGAUAGUAUACGGUCACACCAGGCCUAAUGGGCGCACUUUUUGAAAGUUUU---U ((((((......((((((....))))))(((..((((.((((((...((((((...........)))))).))))))...)))).....)))......))))))......---. ( -30.20) >DroSim_CAF1 4907 114 + 1 UUCGAAUUUCGUUAUCGACUUAUCGAUAGGCAUGGCCAUGUGAUUCAGUAUAUUCGUUUCGAUAUUAUACGGUCACACCAAGCCUAAUGGGCGCACUUUUUGAACGUUUUUUUU ((((((...((((.((((....))))(((((.(((...((((((...(((((.(((...)))...))))).))))))))).)))))...)))).....)))))).......... ( -28.60) >DroEre_CAF1 6608 101 + 1 UUCAAAUUUCU--------UUAUCGAUAGCUGUGGCCAUGUGUUUUAGUAUAUACAUUUGCUUACUAUAUGGUCACACUUGACGAAAGAGGCCCACUUUUUGAAAGUA-----U ((((((..(((--------((.(((.(((.(((((((((((((...((((........)))).)).))))))))))).))).))).))))).......))))))....-----. ( -27.50) >DroYak_CAF1 5012 108 + 1 UUCAAAUUUCUUGAUCGAUUUAUCGAUAGCUGUGGCCAUGUGUUUCAGUAUAUAUGUUUUCUUACUAUACGGUCACACUUGGCCUAAGAGGCGGACUUUUUGAAA-UA-----U ((((((..(((..(((((....))))).(((..(((((.((((..(.(((((((........)).))))).)..)))).))))).....))))))...)))))).-..-----. ( -28.70) >consensus UUCAAAUUUCGUUAUCGACUUAUCGAUAGCCAUGGCCAUGUGUUUCAGUAUAUACGUUUCCAUACUAUACGGUCACACUAGGCCUAAGGGGCGCACUUUUUGAAAGUU_____U ((((((......(((((......)))))(((..((((..((((....(((((.............)))))....))))..)))).....)))......)))))).......... (-14.60 = -16.64 + 2.04)

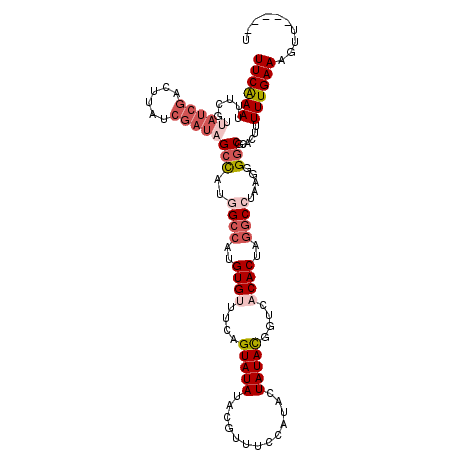
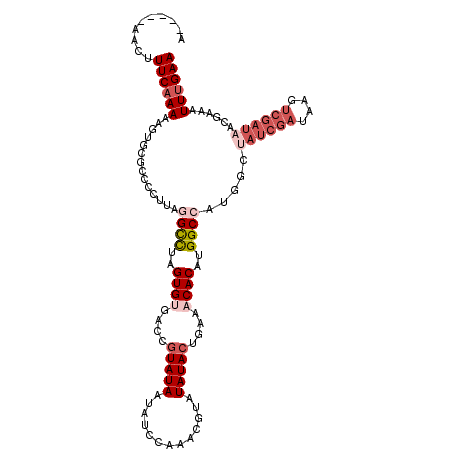
| Location | 16,604,889 – 16,604,999 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.71 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -12.12 |
| Energy contribution | -14.40 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16604889 110 - 20766785 A----GAAGUUUCAAAAAAUCCGCCCCUUGUGCCUAGUGUGACCGUAUAAUAUACAACCGUAUAUACUGAAACACAUGGCCGUGGCUAUCGAUAAGUCGAUAACGAAAUUUGAA .----.(((((((.........(((...((.(((..((((..(.(((((...(((....)))))))).)..))))..))))).)))((((((....))))))..)))))))... ( -27.90) >DroSec_CAF1 4852 111 - 1 A---AAAACUUUCAAAAAGUGCGCCCAUUAGGCCUGGUGUGACCGUAUACUAUCGAAAAGUAUAUACUGAAUCACAUGGCCAUGGCUAUCGAUAGGUCGAUAACGAAAUUUGAA .---......((((((......(((.....((((..((((((..(((((((.......)))))))......))))))))))..)))((((((....))))))......)))))) ( -27.60) >DroSim_CAF1 4907 114 - 1 AAAAAAAACGUUCAAAAAGUGCGCCCAUUAGGCUUGGUGUGACCGUAUAAUAUCGAAACGAAUAUACUGAAUCACAUGGCCAUGCCUAUCGAUAAGUCGAUAACGAAAUUCGAA ........(((.(.....).))).....(((((.(((((((((.(((((...(((...))).))))).)..))))...)))).)))))((((....(((....)))...)))). ( -23.80) >DroEre_CAF1 6608 101 - 1 A-----UACUUUCAAAAAGUGGGCCUCUUUCGUCAAGUGUGACCAUAUAGUAAGCAAAUGUAUAUACUAAAACACAUGGCCACAGCUAUCGAUAA--------AGAAAUUUGAA .-----....((((((.(((.((((......((((....))))....(((((.((....))...)))))........))))...))).((.....--------.))..)))))) ( -17.80) >DroYak_CAF1 5012 108 - 1 A-----UA-UUUCAAAAAGUCCGCCUCUUAGGCCAAGUGUGACCGUAUAGUAAGAAAACAUAUAUACUGAAACACAUGGCCACAGCUAUCGAUAAAUCGAUCAAGAAAUUUGAA .-----..-.((((((...((.........(((((.((((..(.(((((.............))))).)..)))).)))))......(((((....)))))...))..)))))) ( -24.02) >consensus A_____AACUUUCAAAAAGUGCGCCCCUUAGGCCUAGUGUGACCGUAUAAUAUCCAAACGUAUAUACUGAAACACAUGGCCAUGGCUAUCGAUAAGUCGAUAACGAAAUUUGAA ..........((((((..............((((..((((....(((((.............)))))....))))..)))).....((((((....))))))......)))))) (-12.12 = -14.40 + 2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:41 2006