
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,603,519 – 16,603,709 |
| Length | 190 |
| Max. P | 0.999974 |

| Location | 16,603,519 – 16,603,629 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.35 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16603519 110 - 20766785 CAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAAUCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUAAAUU-UCCUACAAAU-UUACAAACAUGAUAUAACUGAAAA .........(((((((((((((((((((.(((....)))..))))))..))))).)))))))).......((((((-(.....))))-)))..................... ( -17.60) >DroSec_CAF1 3511 108 - 1 CAAAACCUUAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUAAAUU-UCCUACAAAU-UUACA-UCAUG-UUUAACUGAAAA ..((((..((((((((((((((((((((.((......))..))))))..))))).)))))))))......((((((-(.....))))-)))..-....)-)))......... ( -18.50) >DroSim_CAF1 3532 108 - 1 CAAAACCUUAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUAAAUU-UCCUACAAAU-UUACA-UCAUG-UUUAACUGAAAA ..((((..((((((((((((((((((((.((......))..))))))..))))).)))))))))......((((((-(.....))))-)))..-....)-)))......... ( -18.50) >DroEre_CAF1 5256 108 - 1 CAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUAAAUU-UCCUAAAAAU-UUACA-UCAUG-UUUAACUGAAAA ..((((...(((((((((((((((((((.((......))..))))))..))))).)))))))).......((((((-(.....))))-)))..-....)-)))......... ( -17.60) >DroYak_CAF1 3667 110 - 1 CAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCUUAAAUGAUACUAAAAUAAAUUUUCCUAAAAAUUUUACG-CCAUG-UUUAACUGAAAA ..((((...(((((((((((((((((((.((......))..))))))..))))).))))))))........(((((((....)))))))....-....)-)))......... ( -16.70) >consensus CAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUAAAUU_UCCUACAAAU_UUACA_UCAUG_UUUAACUGAAAA .........(((((((((((((((((((.((......))..))))))..))))).))))))))................................................. (-16.08 = -16.08 + 0.00)

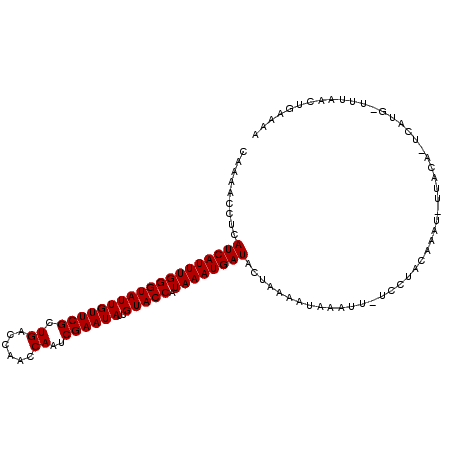
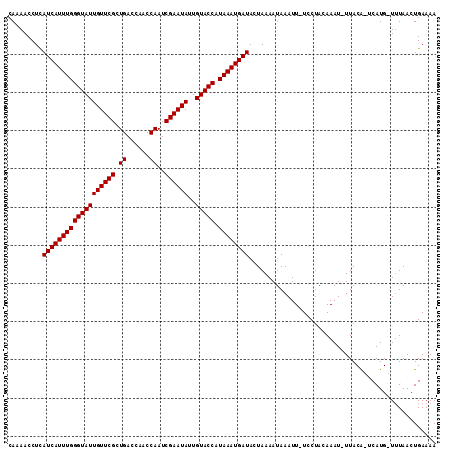
| Location | 16,603,557 – 16,603,669 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.71 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16603557 112 - 20766785 GUACAAUCCAAUUGUUGUACAUGUCCAAAUAGUUUCUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAAUCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUA (((((((......))))))).............................(((((((((((((((((((.(((....)))..))))))..))))).))))))))......... ( -23.20) >DroSec_CAF1 3547 112 - 1 GUACAAUUCAAUUGUUGUACAUGUCCAAAUAGUUCUUAAGCAAAACCUUAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUA (((((((......)))))))............................((((((((((((((((((((.((......))..))))))..))))).)))))))))........ ( -24.20) >DroSim_CAF1 3568 112 - 1 GUACAAUUCAAUUAUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUUAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUA (((((((......)))))))...........(((((.....)))))..((((((((((((((((((((.((......))..))))))..))))).)))))))))........ ( -24.30) >DroEre_CAF1 5292 112 - 1 GUACAAUGACAUUGUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUA (((((((......)))))))...........(((((.....)))))...(((((((((((((((((((.((......))..))))))..))))).))))))))......... ( -23.50) >DroYak_CAF1 3705 112 - 1 GUACAAUCAAAUAGUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCUUAAAUGAUACUAAAAUA (((((((......)))))))...........(((((.....)))))...(((((((((((((((((((.((......))..))))))..))))).))))))))......... ( -23.00) >consensus GUACAAUCCAAUUGUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAAUCGAAUAUUGUACCAUAAAUGAUACUAAAAUA (((((((......))))))).............................(((((((((((((((((((.((......))..))))))..))))).))))))))......... (-23.08 = -22.92 + -0.16)

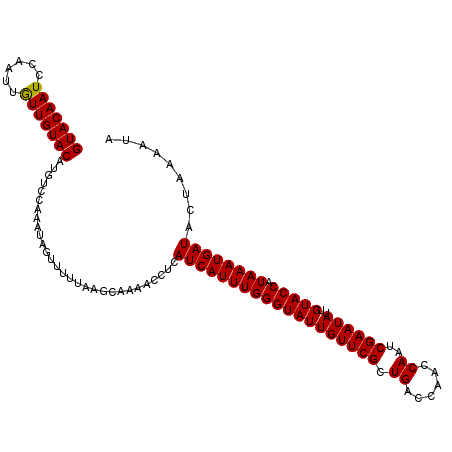
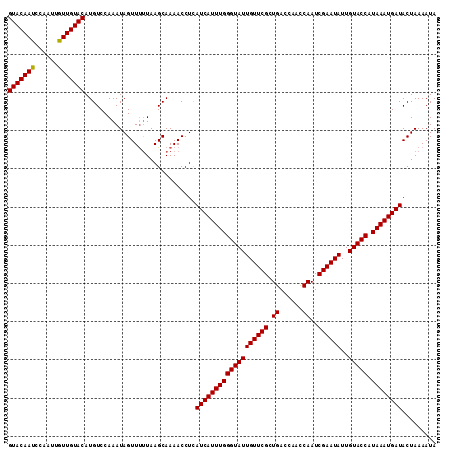
| Location | 16,603,589 – 16,603,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16603589 120 - 20766785 GUUUUCAGUAUCAGAUCCUAAUCAAAACAUUUAAAGAAGUGUACAAUCCAAUUGUUGUACAUGUCCAAAUAGUUUCUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAAUCAA ....(((((...(((..(((.((............)).(((((((((......))))))))).......)))..))).(((((.(((.((.....))))).))))).)))))........ ( -23.20) >DroSec_CAF1 3579 120 - 1 GUUUUCAGUAUCAGAUCCUAAUCAAAACAUUUAAAGAAGUGUACAAUUCAAUUGUUGUACAUGUCCAAAUAGUUCUUAAGCAAAACCUUAUCAUUUGGGUAUUGUUCGCUGACCAACCAA ....(((((..........................((.(((((((((......))))))))).)).............(((((.((((........)))).))))).)))))........ ( -23.00) >DroSim_CAF1 3600 120 - 1 GUUUUCUGUAUCAGAUCCUAAUCAAAACAUUUAAAGAAGUGUACAAUUCAAUUAUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUUAUCAUUUGGGUAUUGUUCGCUGACCAACCAA ..........((((.........(((((((((...((.(((((((((......))))))))).)).)))).)))))..(((((.((((........)))).)))))..))))........ ( -22.70) >DroEre_CAF1 5324 117 - 1 GUUUUCAGUAUCAGAUCCUUAUCAAAACAUUUAA---AGUGUACAAUGACAUUGUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAA ....(((((..............(((((((((..---.(((((((((......)))))))))....)))).)))))..(((((.(((.((.....))))).))))).)))))........ ( -24.00) >DroYak_CAF1 3737 117 - 1 GUUUUUAGUAUCAGAUCCUAAUCAAAACAUUUAA---AGUGUACAAUCAAAUAGUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAA ..........((((.........(((((((((..---.(((((((((......)))))))))....)))).)))))..(((((.(((.((.....))))).)))))..))))........ ( -21.90) >consensus GUUUUCAGUAUCAGAUCCUAAUCAAAACAUUUAAAGAAGUGUACAAUCCAAUUGUUGUACAUGUCCAAAUAGUUUUUAAGCAAAACCUCAUCAUUUGGGUAUUGUUCGCUGACCAACCAA ....(((((..............(((((((((......(((((((((......)))))))))....)))).)))))..(((((.((((........)))).))))).)))))........ (-20.48 = -20.76 + 0.28)

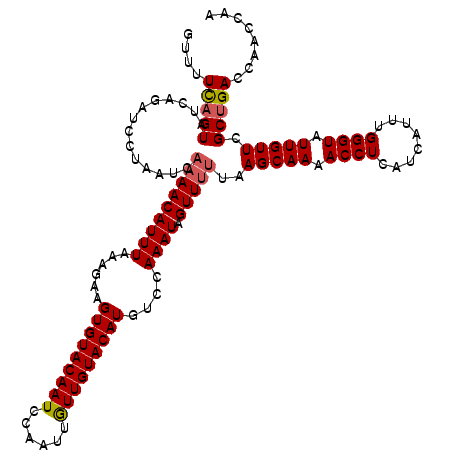
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:40 2006